Establishment of a prognostic signature based on fatty acid metabolism genes in HCC associated with hepatitis B
- PMID: 37957550
- PMCID: PMC10644542
- DOI: 10.1186/s12876-023-03026-5
Establishment of a prognostic signature based on fatty acid metabolism genes in HCC associated with hepatitis B
Abstract
Background: Hepatitis B virus (HBV)-associated hepatocellular carcinoma (HCC) is one of the most common and deadly cancer and often accompanied by varying degrees of liver damage, leading to the dysfunction of fatty acid metabolism (FAM). This study aimed to investigate the relationship between FAM and HBV-associated HCC and identify FAM biomarkers for predicting the prognosis of HBV-associated HCC.
Methods: Gene Set Enrichment Analysis (GSEA) was used to analyze the difference of FAM pathway between paired tumor and adjacent normal tissue samples in 58 HBV-associated HCC patients from the Gene Expression Omnibus (GEO) database. Next, 117 HBV-associated HCC patients from The Cancer Genome Atlas (TCGA) database were analyzed to establish a prognostic signature based on 42 FAM genes. Then, the prognostic signature was validated in an external cohort consisting of 30 HBV-associated HCC patients. Finally, immune infiltration analysis was performed to evaluate the FAM-related immune cells in HBV-associated HCC.
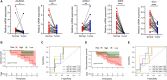
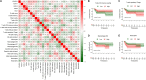
Results: As a result, FAM pathway was clearly downregulated in tumor tissue of HBV-associated HCC, and survival analysis demonstrated that 12 FAM genes were associated with the prognosis of HBV-associated HCC. Lasso-penalized Cox regression analysis identified and established a five-gene signature (ACADVL, ACAT1, ACSL3, ADH4 and ECI1), which showed effective discrimination and prediction for the prognosis of HBV-associated HCC both in the TCGA cohort and the validation cohort. Immune infiltration analysis showed that the high-risk group, identified by FAM signature, of HBV-associated HCC had a higher ratio of Tregs, which was associated with the prognosis.
Conclusions: Collectively, these findings suggest that there is a strong connection between FAM and HBV-associated HCC, indicating a potential therapeutic strategy targeting FAM to block the accumulation of Tregs into the tumor microenvironment of HBV-associated HCC.
Keywords: Fatty acid metabolism; HBV; Hepatocellular carcinoma; Prognosis; Tregs.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

