ESR1 Gene Mutations and Liquid Biopsy in ER-Positive Breast Cancers: A Small Step Forward, a Giant Leap for Personalization of Endocrine Therapy?
- PMID: 37958343
- PMCID: PMC10649433
- DOI: 10.3390/cancers15215169
ESR1 Gene Mutations and Liquid Biopsy in ER-Positive Breast Cancers: A Small Step Forward, a Giant Leap for Personalization of Endocrine Therapy?
Abstract
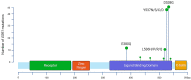
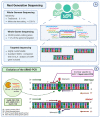
The predominant forms of breast cancer (BC) are hormone receptor-positive (HR+) tumors characterized by the expression of estrogen receptors (ERs) and/or progesterone receptors (PRs). Patients with HR+ tumors can benefit from endocrine therapy (ET). Three types of ET are approved for the treatment of HR+ BCs and include selective ER modulators, aromatase inhibitors, and selective ER downregulators. ET is the mainstay of adjuvant treatment in the early setting and the backbone of the first-line treatment in an advanced setting; however, the emergence of acquired resistance can lead to cancer recurrence or progression. The mechanisms of ET resistance are often related to the occurrence of mutations in the ESR1 gene, which encodes the ER-alpha protein. As ESR1 mutations are hardly detectable at diagnosis but are present in 30% to 40% of advanced BC (ABC) after treatment, the timeline of testing is crucial. To manage this resistance, ESR1 testing has recently been recommended; in ER+ HER2- ABC and circulating cell-free DNA, so-called liquid biopsy appears to be the most convenient way to detect the emergence of ESR1 mutations. Technically, several options exist, including Next Generation Sequencing and ultra-sensitive PCR-based techniques. In this context, personalization of ET through the surveillance of ESR1 mutations in the plasma of HR+ BC patients throughout the disease course represents an innovative way to improve the standard of care.
Keywords: ESR1 gene; breast cancer; endocrine therapy; liquid biopsy.
Conflict of interest statement
Jean-Louis Merlin received honoraria, financial support for meetings, and research grants from Menarini Stemline, Novartis, and Astra Zeneca; Alexandre Harlé received honoraria, financial support for meetings, and research grants from Astra Zeneca and Sophia Genetics; and Vincent Masssard received honoraria, financial support for meetings, and research grants from Menarini Stemline. All other authors declare no conflict of interest.
Figures


References
-
- Burstein H.J., DeMichele A., Somerfield M.R., Henry N.L., for the Biomarker Testing and Endocrine and Targeted Therapy in Metastatic Breast Cancer Expert Panels Testing for ESR1 Mutations to Guide Therapy for Hormone Receptor–Positive, Human Epidermal Growth Factor Receptor 2–Negative Metastatic Breast Cancer: ASCO Guideline Rapid Recommendation Update. JCO. 2023;41:3423–3425. doi: 10.1200/JCO.23.00638. - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

