The BRCA1 c.4096+1G>A Is a Founder Variant Which Originated in Ancient Times
- PMID: 37958491
- PMCID: PMC10648645
- DOI: 10.3390/ijms242115507
The BRCA1 c.4096+1G>A Is a Founder Variant Which Originated in Ancient Times
Abstract
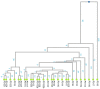
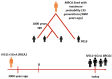
Approximately 30-50% of hereditary breast and ovarian cancer (HBOC) is due to the presence of germline pathogenic variants in the BRCA1 (OMIM 113705) and BRCA2 (OMIM 600185) onco-suppressor genes, which are involved in DNA damage response. Women who carry pathogenic BRCA1 variants are particularly likely to develop breast cancer (BC) and ovarian cancer (OC), with a 45-79 percent and 39-48 percent chance, respectively. The BRCA1 c.4096+1G>A variant has been frequently ascertained in Tuscany, Italy, and it has also been detected in other Italian regions and other countries. Its pathogenetic status has been repeatedly changed from a variant of uncertain significance, to pathogenic, to likely pathogenic. In our study, 48 subjects (38 of whom are carriers) from 27 families were genotyped with the Illumina OncoArray Infinium platform (533,531 SNPs); a 20 Mb region (24.6 cM) around BRCA1, including 4130 SNPs (21 inside BRCA1) was selected for haplotype analysis. We used a phylogenetic method to estimate the time to the most recent common ancestor (MRCA) of BRCA1 c.4096+1G>A founder pathogenic variant. This analysis suggests that the MRCA lived about 155 generations ago-around 3000 years ago.
Keywords: BRCA1; Italy; founder variant; hereditary breast and ovarian cancer; pathogenic variant.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Tung N., Lin N.U., Kidd J., Allen B.A., Singh N., Wenstrup R.J., Hartman A.R., Winer E.P., Garber J.E. Frequency of Germline Mutations in 25 Cancer Susceptibility Genes in a Sequential Series of Patients With Breast Cancer. J. Clin. Oncol. 2016;34:1460–1468. doi: 10.1200/JCO.2015.65.0747. - DOI - PMC - PubMed
-
- Beitsch P.D., Whitworth P.W., Hughes K., Patel R., Rosen B., Compagnoni G., Baron P., Simmons R., Smith L.A., Grady I., et al. Underdiagnosis of Hereditary Breast Cancer: Are Genetic Testing Guidelines a Tool or an Obstacle? J. Clin. Oncol. 2019;37:453–460. doi: 10.1200/JCO.18.01631. - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

