Assessing the Potential Contribution of In Silico Studies in Discovering Drug Candidates That Interact with Various SARS-CoV-2 Receptors
- PMID: 37958503
- PMCID: PMC10647470
- DOI: 10.3390/ijms242115518
Assessing the Potential Contribution of In Silico Studies in Discovering Drug Candidates That Interact with Various SARS-CoV-2 Receptors
Abstract
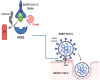
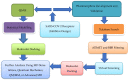
The COVID-19 pandemic has spurred intense research efforts to identify effective treatments for SARS-CoV-2. In silico studies have emerged as a powerful tool in the drug discovery process, particularly in the search for drug candidates that interact with various SARS-CoV-2 receptors. These studies involve the use of computer simulations and computational algorithms to predict the potential interaction of drug candidates with target receptors. The primary receptors targeted by drug candidates include the RNA polymerase, main protease, spike protein, ACE2 receptor, and transmembrane protease serine 2 (TMPRSS2). In silico studies have identified several promising drug candidates, including Remdesivir, Favipiravir, Ribavirin, Ivermectin, Lopinavir/Ritonavir, and Camostat Mesylate, among others. The use of in silico studies offers several advantages, including the ability to screen a large number of drug candidates in a relatively short amount of time, thereby reducing the time and cost involved in traditional drug discovery methods. Additionally, in silico studies allow for the prediction of the binding affinity of the drug candidates to target receptors, providing insight into their potential efficacy. This study is aimed at assessing the useful contributions of the application of computational instruments in the discovery of receptors targeted in SARS-CoV-2. It further highlights some identified advantages and limitations of these studies, thereby revealing some complementary experimental validation to ensure the efficacy and safety of identified drug candidates.
Keywords: ACE2; SARS-CoV-2; TMPRSS2; antiviral activity; drug candidates; in silico studies; molecular docking; molecular dynamics simulations; receptor–ligand complex.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Worldometer COVID-19 Coronavirus Pandemic. [(accessed on 12 October 2023)]. Available online: https://www.worldometers.info/coronavirus/
-
- El-Wahab E.W.A., Eassa S.M., Metwally M., Al-Hraishawi H., Omar S.R. SARS-CoV-2 transmission channels: A review of the literature. MEDICC Rev. 2021;22:51–69. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

