Resistance to 2-Hydroxy-Flutamide in Prostate Cancer Cells Is Associated with the Downregulation of Phosphatidylcholine Biosynthesis and Epigenetic Modifications
- PMID: 37958610
- PMCID: PMC10650717
- DOI: 10.3390/ijms242115626
Resistance to 2-Hydroxy-Flutamide in Prostate Cancer Cells Is Associated with the Downregulation of Phosphatidylcholine Biosynthesis and Epigenetic Modifications
Abstract
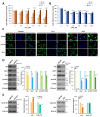
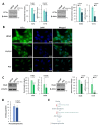
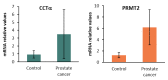
In this study, we examined the metabolic adaptations of a chemoresistant prostate cancer cell line in comparison to a sensitive cell line. We utilized prostate cancer LNCaP cells and subjected them to a stepwise increase in the antiandrogen 2-hydroxy-flutamide (FLU) concentration to generate a FLU-resistant cell line (LN-FLU). These LN-FLU cells displayed characteristics of cancer stem cells, exhibited drug resistance, and showed a significantly reduced expression of Cyclin D1, along with the overexpression of p16, pointing to a proliferation arrest. In comparing the cancer stem-like LN-FLU cells to the LNCaP cells, we observed a decrease in the expression of CTP-choline cytidylyl transferase α (CCTα), as well as a decline in choline kinase, suggesting altogether a downregulation of the phosphatidylcholine biosynthetic pathway. In addition, we found decreased levels of the protein methyl transferase PRMT2 and the upregulation of the histone deacetylase Sirtuin1 (Sirt1). Analysis of the human prostate cancer samples revealed similar results in a population with high expressions of the stem cell markers Oct4 and ABCB1A1. Our findings suggest that the adaptation of prostate cancer cells to antiandrogens could induce reprogramming into stem cells that survive in a low phosphocholine metabolism and cell cycle arrest and display drug resistance.
Keywords: LNCaP cells; antiandrogens; dormant cells; flutamide; phosphatidylcholine metabolism; resistant prostate cancer.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
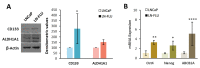

Similar articles
-
Survivin mediates resistance to antiandrogen therapy in prostate cancer.Oncogene. 2005 Apr 7;24(15):2474-82. doi: 10.1038/sj.onc.1208490. Oncogene. 2005. PMID: 15735703
-
Combination of resveratrol and antiandrogen flutamide has synergistic effect on androgen receptor inhibition in prostate cancer cells.Anticancer Res. 2011 Oct;31(10):3323-30. Anticancer Res. 2011. PMID: 21965742
-
Androgenic and antiandrogenic control on epidermal growth factor, epidermal growth factor receptor, and androgen receptor expression in human prostate cancer cell line LNCaP.Prostate. 1995 Jun;26(6):290-8. doi: 10.1002/pros.2990260604. Prostate. 1995. PMID: 7784269
-
Mechanism of action and pure antiandrogenic properties of flutamide.Cancer. 1993 Dec 15;72(12 Suppl):3816-27. doi: 10.1002/1097-0142(19931215)72:12+<3816::aid-cncr2820721711>3.0.co;2-3. Cancer. 1993. PMID: 8252497 Review.
-
Antiandrogens: a summary review of pharmacodynamic properties and tolerability in prostate cancer therapy.Arch Ital Urol Androl. 1999 Dec;71(5):293-302. Arch Ital Urol Androl. 1999. PMID: 10673793 Review.
Cited by
-
Molecular Sentinels: Unveiling the Role of Sirtuins in Prostate Cancer Progression.Int J Mol Sci. 2024 Dec 28;26(1):183. doi: 10.3390/ijms26010183. Int J Mol Sci. 2024. PMID: 39796040 Free PMC article. Review.
-
Special Issue: "Novel Researches and Perspectives on Prostate Cancer".Int J Mol Sci. 2024 Feb 8;25(4):2054. doi: 10.3390/ijms25042054. Int J Mol Sci. 2024. PMID: 38396731 Free PMC article.
-
The role of androgens and global and tissue-specific androgen receptor expression on body composition, exercise adaptation, and performance.Biol Sex Differ. 2025 Apr 23;16(1):28. doi: 10.1186/s13293-025-00707-6. Biol Sex Differ. 2025. PMID: 40269952 Free PMC article. Review.
References
-
- Hanahan D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022;12:31–46. doi: 10.1158/2159-8290.CD-21-1059. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

