Global Proteomics for Identifying the Alteration Pathway of Niemann-Pick Disease Type C Using Hepatic Cell Models
- PMID: 37958627
- PMCID: PMC10648601
- DOI: 10.3390/ijms242115642
Global Proteomics for Identifying the Alteration Pathway of Niemann-Pick Disease Type C Using Hepatic Cell Models
Abstract
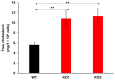
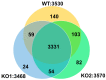
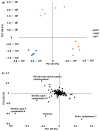
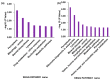
Niemann-Pick disease type C (NPC) is an autosomal recessive disorder with progressive neurodegeneration. Although the causative genes were previously identified, NPC has unclear pathophysiological aspects, and patients with NPC present various symptoms and onset ages. However, various novel biomarkers and metabolic alterations have been investigated; at present, few comprehensive proteomic alterations have been reported in relation to NPC. In this study, we aimed to elucidate proteomic alterations in NPC and perform a global proteomics analysis for NPC model cells. First, we developed two NPC cell models by knocking out NPC1 using CRISPR/Cas9 (KO1 and KO2). Second, we performed a label-free (LF) global proteomics analysis. Using the LF approach, more than 300 proteins, defined as differentially expressed proteins (DEPs), changed in the KO1 and/or KO2 cells, while the two models shared 35 DEPs. As a bioinformatics analysis, the construction of a protein-protein interaction (PPI) network and an enrichment analysis showed that common characteristic pathways such as ferroptosis and mitophagy were identified in the two model cells. There are few reports of the involvement of NPC in ferroptosis, and this study presents ferroptosis as an altered pathway in NPC. On the other hand, many other pathways and DEPs were previously suggested to be associated with NPC, supporting the link between the proteome analyzed here and NPC. Therapeutic research based on these results is expected in the future.
Keywords: Niemann–Pick disease type C; enrichment pathway analysis; global proteomics; knock out; liquid chromatography–electrospray ionization tandem mass spectrometry; model cell.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Maekawa M., Mano N. Identification and Evaluation of Biomarkers for Niemann-Pick Disease Type C Based on Chemical Analysis Techniques. Chromatography. 2020;41:19–29. doi: 10.15583/jpchrom.2020.001. - DOI
MeSH terms
Substances
Grants and funding
- 21K07814/Japan Society for the Promotion of Science
- 19K07075/Japan Society for the Promotion of Science
- Kawano Masanori Memorial Public Interest Incorporated Foundation for Promotion of Pediatrics (Masamitsu Maekawa)
- Japanese Society of Inherited Metabolic Disease/Sanofi LSD Research Grant (Masamitsu Maekawa)
LinkOut - more resources
Full Text Sources
Medical
Research Materials

