A Non-Coding Fc Gamma Receptor Cis-Regulatory Variant within the 1q23 Gene Cluster Is Associated with Plasmodium falciparum Infection in Children Residing in Burkina Faso
- PMID: 37958695
- PMCID: PMC10650193
- DOI: 10.3390/ijms242115711
A Non-Coding Fc Gamma Receptor Cis-Regulatory Variant within the 1q23 Gene Cluster Is Associated with Plasmodium falciparum Infection in Children Residing in Burkina Faso
Abstract
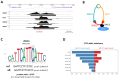
Antibodies play a crucial role in activating protective immunity against malaria by interacting with Fc-gamma receptors (FcγRs). Genetic variations in genes encoding FcγRs can affect immune cell responses to the parasite. In this study, our aim was to investigate whether non-coding variants that regulate FcγR expression could influence the prevalence of Plasmodium falciparum infection. Through bioinformatics approaches, we selected expression quantitative trait loci (eQTL) for FCGR2A, FCGR2B, FCGR2C, FCGR3A, and FCGR3B genes encoding FcγRs (FCGR), in whole blood. We prioritized two regulatory variants, rs2099684 and rs1771575, located in open genomic regions. These variants were identified using RegVar, ImmuNexUT, and transcription factor annotations specific to immune cells. In addition to these, we genotyped the coding variants FCGR2A/rs1801274 and FCGR2B/rs1050501 in 234 individuals from a malaria-endemic area in Burkina Faso. We conducted age and family-based analyses to evaluate associations with the prevalence of malarial infection in both children and adults. The analysis revealed that the regulatory rs1771575-CC genotype was predicted to influence FCGR2B/FCGR2C/FCGR3A transcripts in immune cells and was the sole variant associated with a higher prevalence of malarial infection in children. In conclusion, this study identifies the rs1771575 cis-regulatory variant affecting several FcγRs in myeloid and neutrophil cells and associates it with the inter-individual capacity of children living in Burkina Faso to control malarial infection.
Keywords: Burkina Faso; FCGR ex-pression quantitative trait loci (eQTL); FCGR2A gene polymorphism; FCGR2B gene polymorphism; Fc-gamma receptor (FCGR) gene polymorphism; Fc-gamma receptors/FcγRs; Plasmodium falciparum parasitemia; malaria; regulatory variants.
Conflict of interest statement
The authors declare no conflict of interest and confirm that the funders had no role in the design of the study, the collection, analysis, or interpretation of data, the writing of the manuscript, or in the decision to publish the results.
Figures



References
-
- World Malaria Report. 2022. [(accessed on 4 April 2023)]. Available online: https://www.who.int/teams/global-malaria-programme/reports/world-malaria....
-
- Damena D., Agamah F.E., Kimathi P.O., Kabongo N.E., Girma H., Choga W.T., Golassa L., Chimusa E.R. Insilico Functional Analysis of Genome-Wide Dataset from 17,000 Individuals Identifies Candidate Malaria Resistance Genes Enriched in Malaria Pathogenic Pathways. Front. Genet. 2021;12:676960. doi: 10.3389/fgene.2021.676960. - DOI - PMC - PubMed
-
- Moncunill G., Scholzen A., Mpina M., Nhabomba A., Hounkpatin A.B., Osaba L., Valls R., Campo J.J., Sanz H., Jairoce C., et al. Antigen-stimulated PBMC transcriptional protective signatures for malaria immunization. Sci. Transl. Med. 2020;12:eaay8924. doi: 10.1126/scitranslmed.aay8924. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

