Phylogenetic Analysis and Serological Investigation of Porcine Circovirus Indicates Frequent Infection with Various Subtypes
- PMID: 37958833
- PMCID: PMC10649267
- DOI: 10.3390/ijms242115850
Phylogenetic Analysis and Serological Investigation of Porcine Circovirus Indicates Frequent Infection with Various Subtypes
Abstract
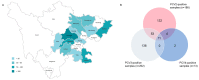
Porcine circoviruses (PCVs) are notorious for triggering severe diseases in pigs and causing serious economic losses to the swine industry. In the present study, we undertook a comprehensive approach for the investigation of PCV prevalence, including the phylogenetic analysis of obtained PCV sequences, the determination of major circulating genotypes and serological screening based on different recombinant Cap proteins with specific immunoreactivity. Epidemiological surveillance data indicate that PCV2d and PCV3a are widely distributed in Southwest China, while PCV4 has only sporadic circulation. Meanwhile, serological investigations showed high PCV2 antibody positivity in collected serum samples (>50%), followed by PCV4 (nearly 50%) and PCV3 (30-35%). The analysis supports different circulation patterns of PCV2, PCV3 and PCV4 and illustrates the PCV2/PCV3 genetic evolution characteristics on a nationwide basis. Taken together, our findings add up to the current understanding of PCV epidemiology and provide new tools and insight for PCV antiviral intervention.
Keywords: Cap; PCV; phylogeny; prevalence; serology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Tischer I., Rasch R., Tochtermann G. Characterization of papovavirus-and picornavirus-like particles in permanent pig kidney cell lines. Zentralbl. Bakteriol. Orig. A. 1974;226:153–167. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

