Genome-Wide Characterization of bHLH Family Genes and Expression Analysis in Response to Osmotic Stress in Betula platyphylla
- PMID: 37960044
- PMCID: PMC10649471
- DOI: 10.3390/plants12213687
Genome-Wide Characterization of bHLH Family Genes and Expression Analysis in Response to Osmotic Stress in Betula platyphylla
Abstract
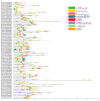
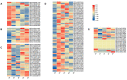
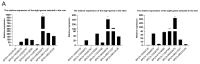
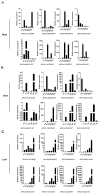
The bHLH family, as a superfamily of transcription factors (TFs), has special functional characteristics in plants and plays a crucial role in a plant's growth and development and helping the plant cope with various stresses. In this study, 128 bHLH family genes were screened in the birch (B. platyphylla) genome using conservative domain scan and blast analysis. These genes are clustered into 21 subfamilies based on the phylogenetic tree construction and are unevenly distributed among the 14 birch chromosomes. In all, 22 segmental duplication pairs with 27 BpbHLH genes were identified. The duplications were distributed on eight chromosomes. Analysis of gene structures and protein motifs revealed intra-group conservation of BpbHLHs. Of the BpbHLH family genes, 16 contain only one intron each. The BPChr14G06667 gene contains the most introns, that is, 19. The cis-elements, which respond to plant hormones, light, defense, and stress, were found on the promoter of BHLH family genes. As per RNA-seq data analysis, under PEG osmotic stress, most BpbHLH genes were differentially expressed, and eight were highly differentially expressed. The qRT-PCR analysis results further indicated that BPChr06G09475 was the gene with the highest expression level in leaves, roots, and stems, and that the expression of these eight genes was higher in leaves than in roots and stems and upregulated in all three tissues under osmotic stress compared to the controls. The above analysis suggests that the BpbHLH family genes have a certain biological effect under drought stress that provides a basis for molecular breeding for stress resistance in birch.
Keywords: RNA-seq; bHLH; birch; drought stress; genome-wide; qRT-PCR.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Han X., Zhao Y., Chen Y., Xu J., Jiang C., Wang X., Zhuo R., Lu M.-Z., Zhang J. Lignin biosynthesis and accumulation in response to abiotic stresses in woody plants. For. Res. 2022;2:9. doi: 10.48130/FR-2022-0009. - DOI
-
- Tikhomirova Tatyana S., Krutovsky Konstantin V., Shestibratov Konstantin A. Molecular Traits for Adaptation to Drought and Salt Stress in Birch, Oak and Poplar Species. Forests. 2022;14:7. doi: 10.3390/f14010007. - DOI
LinkOut - more resources
Full Text Sources
Research Materials

