Quantitative Proteomic Analysis Deciphers the Molecular Mechanism for Endosperm Nuclear Division in Early Rice Seed Development
- PMID: 37960070
- PMCID: PMC10650807
- DOI: 10.3390/plants12213715
Quantitative Proteomic Analysis Deciphers the Molecular Mechanism for Endosperm Nuclear Division in Early Rice Seed Development
Abstract
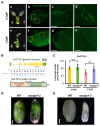
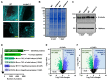
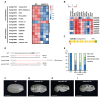
Understanding the molecular mechanisms underlying early seed development is important in improving the grain yield and quality of crop plants. We performed a comparative label-free quantitative proteomic analysis of developing rice seeds for the WT and osctps1-2 mutant, encoding a cytidine triphosphate synthase previously reported as the endospermless 2 (enl2) mutant in rice, harvested at 0 and 1 d after pollination (DAP) to understand the molecular mechanism of early seed development. In total, 5231 proteins were identified, of which 902 changed in abundance between 0 and 1 DAP seeds. Proteins that preferentially accumulated at 1 DAP were involved in DNA replication and pyrimidine biosynthetic pathways. Notably, an increased abundance of OsCTPS1 was observed at 1 DAP; however, no such changes were observed at the transcriptional level. We further observed that the inhibition of phosphorylation increased the stability of this protein. Furthermore, in osctps1-2, minichromosome maintenance (MCM) proteins were significantly reduced compared with those in the WT at 1 DAP, and mutations in OsMCM5 caused defects in seed development. These results highlight the molecular mechanisms underlying early seed development in rice at the post-transcriptional level.
Keywords: CTP synthase; MCM proteins; early seed development; protein stability; proteomic profiling; rice.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
CTP synthase is essential for early endosperm development by regulating nuclei spacing.Plant Biotechnol J. 2021 Nov;19(11):2177-2191. doi: 10.1111/pbi.13644. Epub 2021 Jun 19. Plant Biotechnol J. 2021. PMID: 34058048 Free PMC article.
-
Multi-Omics Approaches Unravel Specific Features of Embryo and Endosperm in Rice Seed Germination.Front Plant Sci. 2022 Jun 9;13:867263. doi: 10.3389/fpls.2022.867263. eCollection 2022. Front Plant Sci. 2022. PMID: 35755645 Free PMC article.
-
Physiological changes and sHSPs genes relative transcription in relation to the acquisition of seed germination during maturation of hybrid rice seed.J Sci Food Agric. 2016 Mar 30;96(5):1764-71. doi: 10.1002/jsfa.7283. Epub 2015 Jun 30. J Sci Food Agric. 2016. PMID: 26031390
-
Use of proteomics to understand seed development in rice.Proteomics. 2013 Jun;13(12-13):1784-800. doi: 10.1002/pmic.201200389. Epub 2013 Apr 23. Proteomics. 2013. PMID: 23483697 Review.
-
Genes and Their Molecular Functions Determining Seed Structure, Components, and Quality of Rice.Rice (N Y). 2022 Mar 18;15(1):18. doi: 10.1186/s12284-022-00562-8. Rice (N Y). 2022. PMID: 35303197 Free PMC article. Review.
Cited by
-
The Emerging Role of Non-Coding RNAs (ncRNAs) in Plant Growth, Development, and Stress Response Signaling.Noncoding RNA. 2024 Feb 7;10(1):13. doi: 10.3390/ncrna10010013. Noncoding RNA. 2024. PMID: 38392968 Free PMC article. Review.
-
Omics-driven advances in the understanding of regulatory landscape of peanut seed development.Front Plant Sci. 2024 May 3;15:1393438. doi: 10.3389/fpls.2024.1393438. eCollection 2024. Front Plant Sci. 2024. PMID: 38766472 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

