Genome-Wide Identification and Characterization of the HAK Gene Family in Quinoa (Chenopodium quinoa Willd.) and Their Expression Profiles under Saline and Alkaline Conditions
- PMID: 37960103
- PMCID: PMC10650088
- DOI: 10.3390/plants12213747
Genome-Wide Identification and Characterization of the HAK Gene Family in Quinoa (Chenopodium quinoa Willd.) and Their Expression Profiles under Saline and Alkaline Conditions
Abstract
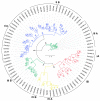
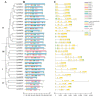
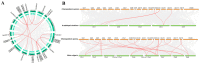
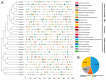
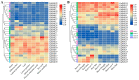
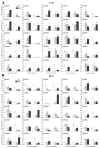
The high-affinity K+ transporter (HAK) family, the most prominent potassium transporter family in plants, which involves K+ transport, plays crucial roles in plant responses to abiotic stresses. However, the HAK gene family remains to be characterized in quinoa (Chenopodium quinoa Willd.). We explored HAKs in quinoa, identifying 30 members (CqHAK1-CqHAK30) in four clusters phylogenetically. Uneven distribution was observed across 18 chromosomes. Furthermore, we investigated the proteins' evolutionary relationships, physicochemical properties, conserved domains and motifs, gene structure, and cis-regulatory elements of the CqHAKs family members. Transcription data analysis showed that CqHAKs have diverse expression patterns among different tissues and in response to abiotic stresses, including drought, heat, low phosphorus, and salt. The expressional changes of CqHAKs in roots were more sensitive in response to abiotic stress than that in shoot apices. Quantitative RT-PCR analysis revealed that under high saline condition, CqHAK1, CqHAK13, CqHAK19, and CqHAK20 were dramatically induced in leaves; under alkaline condition, CqHAK1, CqHAK13, CqHAK19, and CqHAK20 were dramatically induced in leaves, and CqHAK6, CqHAK9, CqHAK13, CqHAK23, and CqHAK29 were significantly induced in roots. Our results establish a foundation for further investigation of the functions of HAKs in quinoa. It is the first study to identify the HAK gene family in quinoa, which provides potential targets for further functional study and contributes to improving the salt and alkali tolerance in quinoa.
Keywords: HAK gene family; alkali stress; expression profile; quinoa; salt stress.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Leigh R.A., Wyn Jones R.G. A hypothesis relating critical potassium concentrations for growth to the distribution and functions of this ion in the plant cell. New Phytol. 1984;97:1–13. doi: 10.1111/j.1469-8137.1984.tb04103.x. - DOI
-
- Hu W., Yang J., Meng Y., Wang Y., Chen B., Zhao W., Oosterhuis D.M., Zhou Z.J. Potassium application affects carbohydrate metabolism in the leaf subtending the cotton (Gossypium hirsutum L.) boll and its relationship with boll biomass. Field Crops Res. 2015;179:120–131. doi: 10.1016/j.fcr.2015.04.017. - DOI
-
- Ye T., Li Y., Zhang J., Hou W., Zhou W., Lu J., Xing Y., Li X. Nitrogen, phosphorus, and potassium fertilization affects the flowering time of rice (Oryza sativa L.) Glob. Ecol. Conserv. 2019;20:e00753. doi: 10.1016/j.gecco.2019.e00753. - DOI
-
- Chambi-Legoas R., Chaix G., Castro V.R., Franco M.P., Tomazello-Filho M.J. Inter-annual effects of potassium/sodium fertilization and water deficit on wood quality of Eucalyptus grandis trees over a full rotation. For. Ecol. Manag. 2021;496:119415. doi: 10.1016/j.foreco.2021.119415. - DOI
-
- Ali M.M., Petropoulos S.A., Selim D.A., Elbagory M., Othman M.M., Omara A.E., Mohamed M.H. Plant growth, yield and quality of potato crop in relation to potassium fertilization. Agronomy. 2021;11:675. doi: 10.3390/agronomy11040675. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

