Systematically and Comprehensively Understanding the Regulation of Cotton Fiber Initiation: A Review
- PMID: 37960127
- PMCID: PMC10648247
- DOI: 10.3390/plants12213771
Systematically and Comprehensively Understanding the Regulation of Cotton Fiber Initiation: A Review
Abstract
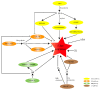
Cotton fibers provide an important source of raw materials for the textile industry worldwide. Cotton fiber is a kind of single cell that differentiates from the epidermis of the ovule and provides a perfect research model for the differentiation and elongation of plant cells. Cotton fiber initiation is the first stage throughout the entire developmental process. The number of fiber cell initials on the seed ovule epidermis decides the final fiber yield. Thus, it is of great significance to clarify the mechanism underlying cotton fiber initiation. Fiber cell initiation is controlled by complex and interrelated regulatory networks. Plant phytohormones, transcription factors, sugar signals, small signal molecules, functional genes, non-coding RNAs, and histone modification play important roles during this process. Here, we not only summarize the different kinds of factors involved in fiber cell initiation but also discuss the mechanisms of these factors that act together to regulate cotton fiber initiation. Our aim is to synthesize a systematic and comprehensive review of different factors during fiber initiation that will provide the basics for further illustrating these mechanisms and offer theoretical guidance for improving fiber yield in future molecular breeding work.
Keywords: cotton; fiber initiation; molecular mechanism; phytohormones; transcription factors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

