Deep Learning Framework for Liver Segmentation from T1-Weighted MRI Images
- PMID: 37960589
- PMCID: PMC10650219
- DOI: 10.3390/s23218890
Deep Learning Framework for Liver Segmentation from T1-Weighted MRI Images
Abstract
The human liver exhibits variable characteristics and anatomical information, which is often ambiguous in radiological images. Machine learning can be of great assistance in automatically segmenting the liver in radiological images, which can be further processed for computer-aided diagnosis. Magnetic resonance imaging (MRI) is preferred by clinicians for liver pathology diagnosis over volumetric abdominal computerized tomography (CT) scans, due to their superior representation of soft tissues. The convenience of Hounsfield unit (HoU) based preprocessing in CT scans is not available in MRI, making automatic segmentation challenging for MR images. This study investigates multiple state-of-the-art segmentation networks for liver segmentation from volumetric MRI images. Here, T1-weighted (in-phase) scans are investigated using expert-labeled liver masks from a public dataset of 20 patients (647 MR slices) from the Combined Healthy Abdominal Organ Segmentation grant challenge (CHAOS). The reason for using T1-weighted images is that it demonstrates brighter fat content, thus providing enhanced images for the segmentation task. Twenty-four different state-of-the-art segmentation networks with varying depths of dense, residual, and inception encoder and decoder backbones were investigated for the task. A novel cascaded network is proposed to segment axial liver slices. The proposed framework outperforms existing approaches reported in the literature for the liver segmentation task (on the same test set) with a dice similarity coefficient (DSC) score and intersect over union (IoU) of 95.15% and 92.10%, respectively.
Keywords: MRI; T1-weighted contrast; automated liver segmentation; deep learning; diagnostic radiology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




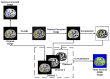
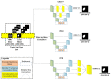


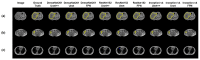

References
-
- Barragán-Montero A., Javaid U., Valdés G., Nguyen D., Desbordes P., Macq B., Willems S., Vandewinckele L., Holmström M., Löfman F., et al. Artificial intelligence and machine learning for medical imaging: A technology review. Phys. Med. 2021;83:242–256. doi: 10.1016/j.ejmp.2021.04.016. - DOI - PMC - PubMed
-
- Chen Z., Song Y., Chang T.-H., Wan X. Generating Radiology Reports via Memory-Driven Transformer. arXiv. 20202010.16056
-
- Tahir A.M., Chowdhury M.E., Khandakar A., Rahman T., Qiblawey Y., Khurshid U., Kiranyaz S., Ibtehaz N., Rahman M.S., Al-Maadeed S., et al. COVID-19 infection localization and severity grading from chest X-ray images. Comput. Biol. Med. 2021;139:105002. doi: 10.1016/j.compbiomed.2021.105002. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

