This is a preprint.
Distinct members of the C. elegans CeMbio reference microbiota exert cryptic virulence and infection protection
- PMID: 37961109
- PMCID: PMC10635080
- DOI: 10.1101/2023.11.02.565327
Distinct members of the C. elegans CeMbio reference microbiota exert cryptic virulence and infection protection
Update in
-
Distinct members of the Caenorhabditis elegans CeMbio reference microbiota exert cryptic virulence that is masked by host defense.Mol Microbiol. 2024 Sep;122(3):387-402. doi: 10.1111/mmi.15258. Epub 2024 Apr 16. Mol Microbiol. 2024. PMID: 38623070 Free PMC article.
Abstract
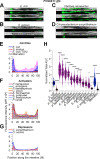
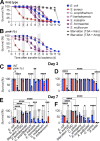
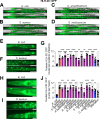
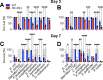
Microbiotas are complex microbial communities that colonize specific niches in the host and provide essential organismal functions that are important in health and disease. A key aspect is the ability of each distinct community member to promote or impair host health, alone or in the context of the community, in hosts with varied levels of immune competence. Understanding such interactions is limited by the complexity and experimental accessibility of current systems and models. Recently, a reference twelve-member microbiota for the model organism C. elegans, known as CeMbio, was defined to aid the dissection of conserved host-microbiota interactions. Understanding the physiological impact of the CeMbio bacteria on C. elegans is in its infancy. Here, we show the differential ability of each CeMbio bacterial species to activate innate immunity through the conserved PMK-1/p38 MAPK, ACh/WNT, and HLH-30/TFEB pathways. Using immunodeficient animals, we uncovered several examples of bacterial 'cryptic' virulence, or virulence that was masked by the host defense response. The ability to activate the PMK-1/p38 pathway did not correlate with bacterial virulence in wild type or immunodeficient animals. In contrast, ten out of twelve species activated HLH-30/TFEB, and most showed virulence towards hlh-30-deficient animals. In addition, we identified Pseudomonas lurida as a pathogen in wild type animals, and Acinetobacter guillouiae as avirulent despite activating all three pathways. Moreover, short pre-exposure to A. guillouiae promoted host survival of infection with P. lurida, which was dependent on PMK-1/p38 MAPK and HLH-30/TFEB. These results suggest that the microbiota of C. elegans is rife with "opportunistic" pathogens, and that HLH-30/TFEB is a fundamental and key host protective factor. Furthermore, they support the idea that bacteria like A. guillouiae evolved the ability to induce host innate immunity to improve host fitness when confronted with pathogens, providing new insights into how colonization order impacts host health.
Figures



Similar articles
-
Distinct members of the Caenorhabditis elegans CeMbio reference microbiota exert cryptic virulence that is masked by host defense.Mol Microbiol. 2024 Sep;122(3):387-402. doi: 10.1111/mmi.15258. Epub 2024 Apr 16. Mol Microbiol. 2024. PMID: 38623070 Free PMC article.
-
CeMbio - The Caenorhabditis elegans Microbiome Resource.G3 (Bethesda). 2020 Sep 2;10(9):3025-3039. doi: 10.1534/g3.120.401309. G3 (Bethesda). 2020. PMID: 32669368 Free PMC article.
-
Association with soil bacteria enhances p38-dependent infection resistance in Caenorhabditis elegans.Infect Immun. 2013 Feb;81(2):514-20. doi: 10.1128/IAI.00653-12. Epub 2012 Dec 10. Infect Immun. 2013. PMID: 23230286 Free PMC article.
-
Autophagy and innate immunity: Insights from invertebrate model organisms.Autophagy. 2018;14(2):233-242. doi: 10.1080/15548627.2017.1389824. Epub 2018 Feb 17. Autophagy. 2018. PMID: 29130360 Free PMC article. Review.
-
Take a Walk to the Wild Side of Caenorhabditis elegans-Pathogen Interactions.Microbiol Mol Biol Rev. 2021 Mar 17;85(2):e00146-20. doi: 10.1128/MMBR.00146-20. Print 2021 May 19. Microbiol Mol Biol Rev. 2021. PMID: 33731489 Free PMC article. Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
