This is a preprint.
The shared genetic architecture and evolution of human language and musical rhythm
- PMID: 37961248
- PMCID: PMC10634981
- DOI: 10.1101/2023.11.01.564908
The shared genetic architecture and evolution of human language and musical rhythm
Update in
-
The shared genetic architecture and evolution of human language and musical rhythm.Nat Hum Behav. 2025 Feb;9(2):376-390. doi: 10.1038/s41562-024-02051-y. Epub 2024 Nov 21. Nat Hum Behav. 2025. PMID: 39572686 Free PMC article.
Abstract
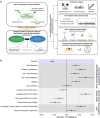
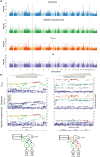
Rhythm and language-related traits are phenotypically correlated, but their genetic overlap is largely unknown. Here, we leveraged two large-scale genome-wide association studies performed to shed light on the shared genetics of rhythm (N=606,825) and dyslexia (N=1,138,870). Our results reveal an intricate shared genetic and neurobiological architecture, and lay groundwork for resolving longstanding debates about the potential co-evolution of human language and musical traits.
Figures


References
-
- Mehr S.A. et al. Science 366, eaax0868 (2019).
-
- Savage P.E. et al. Behavioral and Brain Sciences 44, e59 (2021). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
