This is a preprint.
Senescent and disease-associated microglia are modifiable features of aged brain white matter
- PMID: 37961365
- PMCID: PMC10635389
- DOI: 10.21203/rs.3.rs-3467812/v1
Senescent and disease-associated microglia are modifiable features of aged brain white matter
Abstract
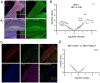
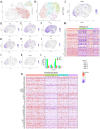
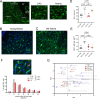
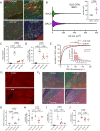
Brain white matter tracts undergo structural and functional changes linked to late-life cognitive decline, but the cellular and molecular contributions to their selective vulnerability are not well defined. In naturally aged mice, we demonstrate that senescent and disease-associated microglia (DAM) phenotypes converge in hippocampus-adjacent white matter. Through gold-standard gene expression and immunolabeling combined with high-dimensional spatial mapping, we identified microglial cell fates in aged white matter characterized by aberrant morphology, microenvironment reorganization, and expression of senescence and DAM markers, including galectin 3 (GAL3/Lgals3), B-cell lymphoma 2 (Bcl2), and cyclin dependent kinase inhibitors, including Cdkn2a/p16ink4a. Pharmacogenetic or pharmacological targeting of p16ink4a or BCL2 reduced white matter GAL3+ DAM abundance and rejuvenated microglial fimbria organization. Our results demonstrate dynamic changes in microglial identity in aged white matter that can be reverted by senotherapeutic intervention to promote homeostatic maintenance in the aged brain.
Keywords: aging; microglia; neuroinflammation; senotherapeutics; white matter.
Conflict of interest statement
CONFLICT OF INTEREST STATEMENT M.J.S., C.M.C., and Mayo Clinic have intellectual property related to this research. This research was reviewed by the Mayo Clinic Conflict of Interest Review Board and was conducted in compliance with Mayo Clinic Conflict of Interest policies. All other authors declare no conflicts.
Figures






References
-
- Papez JW. A proposed mechanism of emotion. 1937. J Neuropsychiatry Clin Neurosci 7, 103–112 (1995). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

