This is a preprint.
Structure of the human heparan-α-glucosaminide N-acetyltransferase (HGSNAT)
- PMID: 37961489
- PMCID: PMC10634761
- DOI: 10.1101/2023.10.23.563672
Structure of the human heparan-α-glucosaminide N-acetyltransferase (HGSNAT)
Update in
-
Structure of the human heparan-α-glucosaminide N-acetyltransferase (HGSNAT).Elife. 2024 Aug 28;13:RP93510. doi: 10.7554/eLife.93510. Elife. 2024. PMID: 39196614 Free PMC article.
Abstract
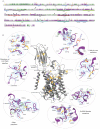
Degradation of heparan sulfate (HS), a glycosaminoglycan (GAG) comprised of repeating units of N-acetylglucosamine and glucuronic acid, begins in the cytosol and is completed in the lysosomes. Acetylation of the terminal non-reducing amino group of a-D-glucosamine of HS is essential for its complete breakdown into monosaccharides and free sulfate. Heparan-a-glucosaminide N-acetyltransferase (HGSNAT), a resident of the lysosomal membrane, catalyzes this essential acetylation reaction by accepting and transferring the acetyl group from cytosolic acetyl-CoA to terminal a-D-glucosamine of HS in the lysosomal lumen. Mutation-induced dysfunction in HGSNAT causes abnormal accumulation of HS within the lysosomes and leads to an autosomal recessive neurodegenerative lysosomal storage disorder called mucopolysaccharidosis IIIC (MPS IIIC). There are no approved drugs or treatment strategies to cure or manage the symptoms of, MPS IIIC. Here, we use cryo-electron microscopy (cryo-EM) to determine a high-resolution structure of the HGSNAT-acetyl-CoA complex, the first step in HGSNAT catalyzed acetyltransferase reaction. In addition, we map the known MPS IIIC mutations onto the structure and elucidate the molecular basis for mutation-induced HGSNAT dysfunction.
Conflict of interest statement
Conflict of interest The authors declare no conflict of interest.
Figures



Similar articles
-
Structure of the human heparan-α-glucosaminide N-acetyltransferase (HGSNAT).Elife. 2024 Aug 28;13:RP93510. doi: 10.7554/eLife.93510. Elife. 2024. PMID: 39196614 Free PMC article.
-
Analysis of the biogenesis of heparan sulfate acetyl-CoA:alpha-glucosaminide N-acetyltransferase provides insights into the mechanism underlying its complete deficiency in mucopolysaccharidosis IIIC.J Biol Chem. 2010 Oct 8;285(41):31233-42. doi: 10.1074/jbc.M110.141150. Epub 2010 Jul 22. J Biol Chem. 2010. PMID: 20650889 Free PMC article.
-
Novel Direct Assay for Acetyl-CoA:α-Glucosaminide N-Acetyltransferase Using BODIPY-Glucosamine as a Substrate.JIMD Rep. 2015 Oct 23;28:11-8. doi: 10.1007/8904_2015_501. Online ahead of print. JIMD Rep. 2015. PMID: 26493749 Free PMC article.
-
Sanfilippo syndrome type C: mutation spectrum in the heparan sulfate acetyl-CoA: alpha-glucosaminide N-acetyltransferase (HGSNAT) gene.Hum Mutat. 2009 Jun;30(6):918-25. doi: 10.1002/humu.20986. Hum Mutat. 2009. PMID: 19479962 Review.
-
Glycosaminoglycans and mucopolysaccharidosis type III.Front Biosci (Landmark Ed). 2016 Jun 1;21(7):1393-409. doi: 10.2741/4463. Front Biosci (Landmark Ed). 2016. PMID: 27100513 Review.
References
-
- Ausseil J, Landry K, Seyrantepe V, Trudel S, Mazur A, Lapointe F, Pshezhetsky AV (2006) An acetylated 120-kDa lysosomal transmembrane protein is absent from mucopolysaccharidosis IIIC fibroblasts: a candidate molecule for MPS IIIC. Mol Genet Metab 87: 22–31 - PubMed
-
- Bame KJ, Rome LH (1985) Acetyl coenzyme A: alpha-glucosaminide N-acetyltransferase. Evidence for a transmembrane acetylation mechanism. J Biol Chem 260: 11293–11299 - PubMed
-
- Bame KJ, Rome LH (1986a) Acetyl-coenzyme A:alpha-glucosaminide N-acetyltransferase. Evidence for an active site histidine residue. J Biol Chem 261: 10127–10132 - PubMed
-
- Bame KJ, Rome LH (1986b) Genetic evidence for transmembrane acetylation by lysosomes. Science 233: 1087–1089 - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
