This is a preprint.
Aryl hydrocarbon receptor restricts axon regeneration of DRG neurons in response to injury
- PMID: 37961567
- PMCID: PMC10635160
- DOI: 10.1101/2023.11.04.565649
Aryl hydrocarbon receptor restricts axon regeneration of DRG neurons in response to injury
Abstract
Injured neurons sense environmental cues to balance neural protection and axon regeneration, but the mechanisms are unclear. Here, we unveil aryl hydrocarbon receptor (AhR), a ligand-activated bHLH-PAS transcription factor, as a molecular sensor and key regulator of acute stress response at the expense of axon regeneration. We demonstrate responsiveness of DRG sensory neurons to AhR signaling, which functions to inhibit axon regeneration. Conditional Ahr deletion in neurons accelerates axon regeneration after sciatic nerve injury. Ahr deletion partially mimics the conditioning lesion in priming DRG to initiate axonogenesis gene programs; upon peripheral axotomy, Ahr ablation suppresses inflammation and stress signaling while augmenting pro-growth pathways. Moreover, comparative transcriptomics revealed signaling interactions between AhR and HIF-1α, two structurally related bHLH-PAS α units that share the dimerization partner Arnt/HIF-1β. Functional assays showed that the growth advantage of AhR-deficient DRG neurons requires HIF-1α; but in the absence of Arnt, DRG neurons can still mount a regenerative response. We further unveil a link between bHLH-PAS transcription factors and DNA hydroxymethylation in response to peripheral axotomy, while RNA-seq of DRG neurons and neuronal single cell RNA-seq analysis revealed a link of AhR regulon to RNA regulation and integrated stress response (ISR). Altogether, AhR activation favors stress coping and inflammation at the expense of axon regeneration; targeting AhR has the potential to enhance nerve repair.
Keywords: Arnt; Aryl hydrocarbon receptor (AhR); Axon regeneration; Conditioning lesion of DRG; DNA hydroxymethylation; Hypoxia-inducible factor (HIF); Integrated stress response (ISR); Spinal cord injury.
Conflict of interest statement
Competing Interests Statement: All authors declare no competing interests.
Figures
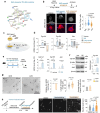
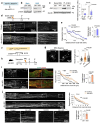
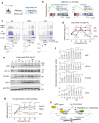

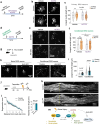

References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
