Adenovirus E1A binding to DCAF10 targets proteasomal degradation of RUVBL1/2 AAA+ ATPases required for quaternary assembly of multiprotein machines, innate immunity, and responses to metabolic stress
- PMID: 37962355
- PMCID: PMC10734532
- DOI: 10.1128/jvi.00993-23
Adenovirus E1A binding to DCAF10 targets proteasomal degradation of RUVBL1/2 AAA+ ATPases required for quaternary assembly of multiprotein machines, innate immunity, and responses to metabolic stress
Abstract
Inactivation of EP300/CREBB paralogous cellular lysine acetyltransferases (KATs) during the early phase of infection is a consistent feature of DNA viruses. The cell responds by stabilizing transcription factor IRF3 which activates transcription of scores of interferon-stimulated genes (ISGs), inhibiting viral replication. Human respiratory adenoviruses counter this by assembling a CUL4-based ubiquitin ligase complex that polyubiquitinylates RUVBL1 and 2 inducing their proteasomal degradation. This inhibits accumulation of active IRF3 and the expression of anti-viral ISGs, allowing replication of the respiratory HAdVs in the face of inhibition of EP300/CBEBBP KAT activity by the N-terminal region of E1A.
Keywords: CBP; CRL4; DCAF10; E1A; IRF3; P300; adenovirus; innate immunity; virology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

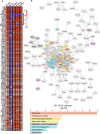
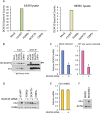
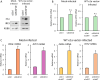
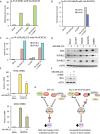
Similar articles
-
Suppression of Type I Interferon Signaling by E1A via RuvBL1/Pontin.J Virol. 2017 Mar 29;91(8):e02484-16. doi: 10.1128/JVI.02484-16. Print 2017 Apr 15. J Virol. 2017. PMID: 28122980 Free PMC article.
-
Adenovirus E1A inhibits SCF(Fbw7) ubiquitin ligase.J Biol Chem. 2009 Oct 9;284(41):27766-27779. doi: 10.1074/jbc.M109.006809. Epub 2009 Aug 13. J Biol Chem. 2009. PMID: 19679664 Free PMC article.
-
Transformation by E1A oncoprotein involves ubiquitin-mediated proteolysis of the neuronal and tumor repressor REST in the nucleus.J Virol. 2012 May;86(10):5594-602. doi: 10.1128/JVI.06811-11. Epub 2012 Mar 14. J Virol. 2012. PMID: 22419809 Free PMC article.
-
Modulation of oncogenic transformation by the human adenovirus E1A C-terminal region.Curr Top Microbiol Immunol. 2004;273:139-61. doi: 10.1007/978-3-662-05599-1_5. Curr Top Microbiol Immunol. 2004. PMID: 14674601 Review.
-
Adenoviral strategies to overcome innate cellular responses to infection.FEBS Lett. 2019 Dec;593(24):3484-3495. doi: 10.1002/1873-3468.13680. Epub 2019 Nov 26. FEBS Lett. 2019. PMID: 31721176 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

