Genetic and functional enrichments associated with Enterococcus faecalis isolated from the urinary tract
- PMID: 37962362
- PMCID: PMC10746210
- DOI: 10.1128/mbio.02515-23
Genetic and functional enrichments associated with Enterococcus faecalis isolated from the urinary tract
Abstract
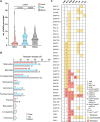
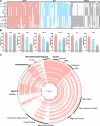
Urinary tract infection (UTI) is a global health issue that imposes a substantial burden on healthcare systems. Women are disproportionately affected by UTI, with >60% of women experiencing at least one UTI in their lifetime. UTIs can recur, particularly in postmenopausal women, leading to diminished quality of life and potentially life-threatening complications. Understanding how pathogens colonize and survive in the urinary tract is necessary to identify new therapeutic targets that are urgently needed due to rising rates of antimicrobial resistance. How Enterococcus faecalis, a bacterium commonly associated with UTI, adapts to the urinary tract remains understudied. Here, we generated a collection of high-quality closed genome assemblies of clinical urinary E. faecalis isolated from the urine of postmenopausal women that we used alongside detailed clinical metadata to perform a robust comparative genomic investigation of genetic factors that may be involved in E. faecalis survival in the urinary tract.
Keywords: Enterococcus; adaptation; genomics; host-microbe interactions; hybrid assembly; urinary tract infection.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
MeSH terms
Substances
Grants and funding
- R01 AI116610/AI/NIAID NIH HHS/United States
- R01 DK131267/DK/NIDDK NIH HHS/United States
- AT-2030-20200401/Welch Foundation (The Welch Foundation)
- R01AI116610/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- 1R01DK131267-01/HHS | NIH | National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK)
LinkOut - more resources
Full Text Sources
Medical
