Propagation of PrPSc in mice reveals impact of aggregate composition on prion disease pathogenesis
- PMID: 37964018
- PMCID: PMC10645910
- DOI: 10.1038/s42003-023-05541-3
Propagation of PrPSc in mice reveals impact of aggregate composition on prion disease pathogenesis
Abstract
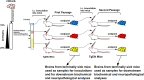
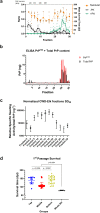
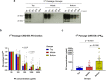
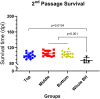
Infectious prions consist of PrPSc, a misfolded, aggregation-prone isoform of the host's prion protein. PrPSc assemblies encode distinct biochemical and biological properties. They harbor a specific profile of PrPSc species, from small oligomers to fibrils in different ratios, where the highest infectivity aligns with oligomeric particles. To investigate the impact of PrPSc aggregate complexity on prion propagation, biochemical properties, and disease pathogenesis, we fractionated elk prions by sedimentation velocity centrifugation, followed by sub-passages of individual fractions in cervidized mice. Upon first passage, different fractions generated PrPSc with distinct biochemical, biophysical, and neuropathological profiles. Notably, low or high molecular weight PrPSc aggregates caused different clinical signs of hyperexcitability or lethargy, respectively, which were retained over passage, whereas other properties converged. Our findings suggest that PrPSc quaternary structure determines an initial selection of a specific replication environment, resulting in transmissible features that are independent of PrPSc biochemical and biophysical properties.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interest.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

