Hyperspectral imaging benchmark based on machine learning for intraoperative brain tumour detection
- PMID: 37964078
- PMCID: PMC10646050
- DOI: 10.1038/s41698-023-00475-9
Hyperspectral imaging benchmark based on machine learning for intraoperative brain tumour detection
Abstract
Brain surgery is one of the most common and effective treatments for brain tumour. However, neurosurgeons face the challenge of determining the boundaries of the tumour to achieve maximum resection, while avoiding damage to normal tissue that may cause neurological sequelae to patients. Hyperspectral (HS) imaging (HSI) has shown remarkable results as a diagnostic tool for tumour detection in different medical applications. In this work, we demonstrate, with a robust k-fold cross-validation approach, that HSI combined with the proposed processing framework is a promising intraoperative tool for in-vivo identification and delineation of brain tumours, including both primary (high-grade and low-grade) and secondary tumours. Analysis of the in-vivo brain database, consisting of 61 HS images from 34 different patients, achieve a highest median macro F1-Score result of 70.2 ± 7.9% on the test set using both spectral and spatial information. Here, we provide a benchmark based on machine learning for further developments in the field of in-vivo brain tumour detection and delineation using hyperspectral imaging to be used as a real-time decision support tool during neurosurgical workflows.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
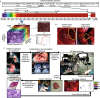






Similar articles
-
Spatio-spectral classification of hyperspectral images for brain cancer detection during surgical operations.PLoS One. 2018 Mar 19;13(3):e0193721. doi: 10.1371/journal.pone.0193721. eCollection 2018. PLoS One. 2018. PMID: 29554126 Free PMC article.
-
Brain tissue classification in hyperspectral images using multistage diffusion features and transformer.J Microsc. 2024 Nov 19. doi: 10.1111/jmi.13372. Online ahead of print. J Microsc. 2024. PMID: 39563208
-
An Intraoperative Visualization System Using Hyperspectral Imaging to Aid in Brain Tumor Delineation.Sensors (Basel). 2018 Feb 1;18(2):430. doi: 10.3390/s18020430. Sensors (Basel). 2018. PMID: 29389893 Free PMC article.
-
Hyperspectral Imaging in Brain Tumor Surgery-Evidence of Machine Learning-Based Performance.World Neurosurg. 2023 Jul;175:e614-e635. doi: 10.1016/j.wneu.2023.03.149. Epub 2023 Apr 6. World Neurosurg. 2023. PMID: 37030483
-
Machine learning techniques for analysis of hyperspectral images to determine quality of food products: A review.Curr Res Food Sci. 2021 Feb 3;4:28-44. doi: 10.1016/j.crfs.2021.01.002. eCollection 2021. Curr Res Food Sci. 2021. PMID: 33659896 Free PMC article. Review.
Cited by
-
Calibration-Jitter: Augmentation of hyperspectral data for improved surgical scene segmentation.Healthc Technol Lett. 2024 Nov 29;11(6):345-354. doi: 10.1049/htl2.12102. eCollection 2024 Dec. Healthc Technol Lett. 2024. PMID: 39720743 Free PMC article.
-
RGB to hyperspectral: Spectral reconstruction for enhanced surgical imaging.Healthc Technol Lett. 2024 Nov 25;11(6):307-317. doi: 10.1049/htl2.12098. eCollection 2024 Dec. Healthc Technol Lett. 2024. PMID: 39720751 Free PMC article.
-
Noninvasive Oral Hyperspectral Imaging-Driven Digital Diagnosis of Heart Failure With Preserved Ejection Fraction: Model Development and Validation Study.J Med Internet Res. 2025 Jan 7;27:e67256. doi: 10.2196/67256. J Med Internet Res. 2025. PMID: 39773415 Free PMC article.
-
Adaptive image encryption approach using an enhanced swarm intelligence algorithm.Sci Rep. 2025 Mar 19;15(1):9476. doi: 10.1038/s41598-025-86569-9. Sci Rep. 2025. PMID: 40108167 Free PMC article.
-
Multimodal imaging platform for enhanced tumor resection in neurosurgery: integrating hyperspectral and pCLE technologies.Int J Comput Assist Radiol Surg. 2025 Jun;20(6):1087-1096. doi: 10.1007/s11548-025-03340-1. Epub 2025 Apr 3. Int J Comput Assist Radiol Surg. 2025. PMID: 40180672 Free PMC article.
References
-
- International Association of Cancer Registries (IACR). Cancer Today. GLOBOCAN 2020.https://gco.iarc.fr/today/home (2021).
-
- International Association of Cancer Registries (IACR). Cancer Tomorrow. GLOBOCAN 2020. https://gco.iarc.fr/tomorrow/en (2021).
-
- National Institute for Health and Care Excellence. Brain Tumours (primary) and Brain Metastases in Adults (NG99). (2018). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

