Characterisation of a novel [18F]FDG brain PET database and combination with a second database for optimising detection of focal abnormalities, using focal cortical dysplasia as an example
- PMID: 37964137
- PMCID: PMC10645721
- DOI: 10.1186/s13550-023-01023-z
Characterisation of a novel [18F]FDG brain PET database and combination with a second database for optimising detection of focal abnormalities, using focal cortical dysplasia as an example
Abstract
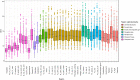
Background: Brain [18F]FDG PET is used clinically mainly in the presurgical evaluation for epilepsy surgery and in the differential diagnosis of neurodegenerative disorders. While scans are usually interpreted visually on an individual basis, comparison against normative cohorts allows statistical assessment of abnormalities and potentially higher sensitivity for detecting abnormalities. Little work has been done on out-of-sample databases (acquired differently to the patient data). Combination of different databases would potentially allow better power and discrimination. We fully characterised an unpublished healthy control brain [18F]FDG PET database (Marseille, n = 60, ages 21-78 years) and compared it to another publicly available database (MRXFDG, n = 37, ages 23-65 years). We measured and then harmonised spatial resolution and global values. A collection of patient scans (n = 34, 13-48 years) with histologically confirmed focal cortical dysplasias (FCDs) obtained on three generations of scanners was used to estimate abnormality detection rates using standard software (statistical parametric mapping, SPM12).
Results: Regional SUVs showed similar patterns, but global values and resolutions were different as expected. Detection rates for the FCDs were 50% for comparison with the Marseille database and 53% for MRXFDG. Simply combining both databases worsened the detection rate to 41%. After harmonisation of spatial resolution, using a full factorial design matrix to accommodate global differences, and leaving out controls older than 60 years, we achieved detection rates of up to 71% for both databases combined. Detection rates were similar across the three scanner types used for patients, and high for patients whose MRI had been normal (n = 10/11).
Conclusions: As expected, global and regional data characteristics are database specific. However, our work shows the value of increasing database size and suggests ways in which database differences can be overcome. This may inform analysis via traditional statistics or machine learning, and clinical implementation.
Keywords: Anomaly detection; Harmonisation; Interscanner differences; Smoothness; Statistical parametric mapping (SPM).
© 2023. The Author(s).
Conflict of interest statement
Academic licences for the Hammers Atlas Database are freely available via
Figures




References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

