Genomic and phenotypic comparison of polyhydroxyalkanoates producing strains of genus Caldimonas/ Schlegelella
- PMID: 37965057
- PMCID: PMC10641440
- DOI: 10.1016/j.csbj.2023.10.051
Genomic and phenotypic comparison of polyhydroxyalkanoates producing strains of genus Caldimonas/ Schlegelella
Abstract
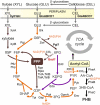
Polyhydroxyalkanoates (PHAs) have emerged as an environmentally friendly alternative to conventional polyesters. In this study, we present a comprehensive analysis of the genomic and phenotypic characteristics of three non-model thermophilic bacteria known for their ability to produce PHAs: Schlegelella aquatica LMG 23380T, Caldimonas thermodepolymerans DSM 15264, and C. thermodepolymerans LMG 21645 and the results were compared with the type strain C. thermodepolymerans DSM 15344T. We have assembled the first complete genomes of these three bacteria and performed the structural and functional annotation. This analysis has provided valuable insights into the biosynthesis of PHAs and has allowed us to propose a comprehensive scheme of carbohydrate metabolism in the studied bacteria. Through phylogenomic analysis, we have confirmed the synonymity between Caldimonas and Schlegelella genera, and further demonstrated that S. aquatica and S. koreensis, currently classified as orphan species, belong to the Caldimonas genus.
Keywords: Caldimonas; DSM 15264; DSM 15344T; LMG 21645; LMG 23380T; Next-Generation Industrial Biotechnology; PHAs; Schlegelella; de novo assembly.
© 2023 The Authors. Published by Elsevier B.V. on behalf of Research Network of Computational and Structural Biotechnology.
Conflict of interest statement
None.
Figures




References
-
- Alcock B.P., Raphenya A.R., Lau T.T.Y., Tsang K.K., Bouchard M., Edalatmand A., Huynh W., Nguyen A.L.V., Cheng A.A., Liu S., Min S.Y., Miroshnichenko A., Tran H.K., Werfalli R.E., Nasir J.A., Oloni M., Speicher D.J., Florescu A., Singh B., Faltyn M., Hernandez-Koutoucheva A., Sharma A.N., Bordeleau E., Pawlowski A.C., Zubyk H.L., Dooley D., Griffiths E., Maguire F., Winsor G.L., Beiko R.G., Brinkman F.S.L., Hsiao W.W.L., Domselaar G.V., McArthur A.G. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020;48:D517–D525. doi: 10.1093/nar/gkz935. - DOI - PMC - PubMed
-
- Berks B.C., Palmer T., Sargent F. Tat Protein Translocat Pathw its role Microb Physiol. 2003:187–254. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
