FXYD3 functionally demarcates an ancestral breast cancer stem cell subpopulation with features of drug-tolerant persisters
- PMID: 37966117
- PMCID: PMC10645391
- DOI: 10.1172/JCI166666
FXYD3 functionally demarcates an ancestral breast cancer stem cell subpopulation with features of drug-tolerant persisters
Abstract
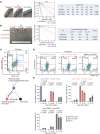
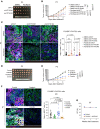
The heterogeneity of cancer stem cells (CSCs) within tumors presents a challenge in therapeutic targeting. To decipher the cellular plasticity that fuels phenotypic heterogeneity, we undertook single-cell transcriptomics analysis in triple-negative breast cancer (TNBC) to identify subpopulations in CSCs. We found a subpopulation of CSCs with ancestral features that is marked by FXYD domain-containing ion transport regulator 3 (FXYD3), a component of the Na+/K+ pump. Accordingly, FXYD3+ CSCs evolve and proliferate, while displaying traits of alveolar progenitors that are normally induced during pregnancy. Clinically, FXYD3+ CSCs were persistent during neoadjuvant chemotherapy, hence linking them to drug-tolerant persisters (DTPs) and identifying them as crucial therapeutic targets. Importantly, FXYD3+ CSCs were sensitive to senolytic Na+/K+ pump inhibitors, such as cardiac glycosides. Together, our data indicate that FXYD3+ CSCs with ancestral features are drivers of plasticity and chemoresistance in TNBC. Targeting the Na+/K+ pump could be an effective strategy to eliminate CSCs with ancestral and DTP features that could improve TNBC prognosis.
Keywords: Breast cancer; Drug therapy; Oncology; Stem cells.
Figures







References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

