A laboratory framework for ongoing optimization of amplification-based genomic surveillance programs
- PMID: 37966271
- PMCID: PMC10715188
- DOI: 10.1128/spectrum.02202-23
A laboratory framework for ongoing optimization of amplification-based genomic surveillance programs
Abstract
This study provides a laboratory framework to ensure ongoing relevance and performance of amplification-based whole genome sequencing to strengthen public health surveillance during extended outbreaks or pandemics. The framework integrates regular reviews of the performance of a genomic surveillance system and highlights the importance of ongoing monitoring and the identification and implementation of improvements to whole genome sequencing methods to enhance public health responses to pathogen outbreaks.
Keywords: genomic surveillance; public health; viral sequencing; whole genome sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


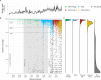
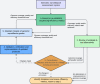
References
-
- Brito AF, Semenova E, Dudas G, Hassler GW, Kalinich CC, Kraemer MUG, Ho J, Tegally H, Githinji G, Agoti CN, et al. . 2021. Global disparities in SARS-CoV-2 genomic surveillance. Cold Spring Harbor Laboratory.
-
- ARTICNetwork . 2020. SARS-CoV-2 ARTIC V3 Illumina library construction and sequencing protocol V.4. Protocols.Io
-
- Eden J-S, Rockett R, Carter I, Rahman H, de Ligt J, Hadfield J, Storey M, Ren X, Tulloch R, Basile K, Wells J, Byun R, Gilroy N, O’Sullivan MV, Sintchenko V, Chen SC, Maddocks S, Sorrell TC, Holmes EC, Dwyer DE, Kok J, 2019-nCoV Study Group . 2020. An emergent clade of SARS-CoV-2 linked to returned travellers from Iran. Virus Evol 6:veaa027. doi:10.1093/ve/veaa027 - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

