Super-enhancer driven SOX2 promotes tumor formation by chromatin re-organization in nasopharyngeal carcinoma
- PMID: 37967508
- PMCID: PMC10679863
- DOI: 10.1016/j.ebiom.2023.104870
Super-enhancer driven SOX2 promotes tumor formation by chromatin re-organization in nasopharyngeal carcinoma
Abstract
Background: Nasopharyngeal carcinoma (NPC) is a malignant head and neck cancer with a high incidence in Southern China and Southeast Asia. Patients with remote metastasis and recurrent NPC have poor prognosis. Thus, a better understanding of NPC pathogenesis may identify novel therapies to address the unmet clinical needs.
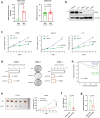
Methods: H3K27ac ChIP-seq and HiChIP was applied to understand the enhancer landscapes and the chromosome interactions. Whole genome sequencing was conducted to analyze the relationship between genomic variations and epigenetic dysregulation. CRISPRi and JQ1 treatment were used to evaluate the transcriptional regulation of SOX2 SEs. Colony formation assay, survival analysis and in vivo subcutaneous patient-derived xenograft assays were applied to explore the function and clinical relevance of SOX2 in NPC.
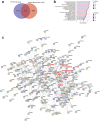
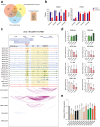
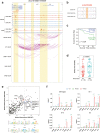
Findings: We globally mapped the enhancer landscapes and generated NPC enhancer connectomes, linking NPC specific enhancers and SEs. We found five overlapped genes, including SOX2, among super-enhancer regulated genes, survival related genes and NPC essential genes. The mRNA expression of SOX2 was repressed when applying CRISPRi targeting different SOX2 SEs or JQ1 treatment. Next, we identified a genetic variation (Chr3:181422197, G > A) in SOX2 SE which is correlated with higher expression of SOX2 and poor survival. In addition, SOX2 was highly expressed in NPC and is correlated with short survival in patients with NPC. Knock-down of SOX2 suppressed tumor growth in vitro and in vivo.
Interpretation: Our study demonstrated the super-enhancer landscape with chromosome interactions and identified super-enhancer driven SOX2 promotes tumorigenesis, suggesting that SOX2 is a potential therapeutic target for patients with NPC.
Funding: A full list of funding bodies that contributed to this study can be found in the Acknowledgements section.
Keywords: Chromatin re-organization; Enhancer connectomes; Genetic variation; Nasopharyngeal carcinoma; Super-enhancers.
Copyright © 2023 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests All authors declare no potential conflicts of interest.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

