Silencing of genes by promoter hypermethylation shapes tumor microenvironment and resistance to immunotherapy in clear-cell renal cell carcinomas
- PMID: 37967556
- PMCID: PMC10694769
- DOI: 10.1016/j.xcrm.2023.101287
Silencing of genes by promoter hypermethylation shapes tumor microenvironment and resistance to immunotherapy in clear-cell renal cell carcinomas
Abstract
The efficacy of immune checkpoint inhibitors varies in clear-cell renal cell carcinoma (ccRCC), with notable primary resistance among patients. Here, we integrate epigenetic (DNA methylation) and transcriptome data to identify a ccRCC subtype characterized by cancer-specific promoter hypermethylation and epigenetic silencing of Polycomb targets. We develop and validate an index of methylation-based epigenetic silencing (iMES) that predicts primary resistance to immune checkpoint inhibition (ICI) in the BIONIKK trial. High iMES is associated with VEGF pathway silencing, endothelial cell depletion, immune activation/suppression, EZH2 activation, BAP1/SETD2 deficiency, and resistance to ICI. Combination therapy with hypomethylating agents or tyrosine kinase inhibitors may benefit patients with high iMES. Intriguingly, tumors with low iMES exhibit increased endothelial cells and improved ICI response, suggesting the importance of angiogenesis in ICI treatment. We also develop a transcriptome-based analogous system for extended applicability of iMES. Our study underscores the interplay between epigenetic alterations and tumor microenvironment in determining immunotherapy response.
Keywords: DNA methylation; angiogenesis; biomarker; clear-cell renal cell carcinoma; epigenetic silencing; immune checkpoint inhibitors; tumor microenvironment.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures

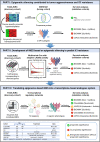




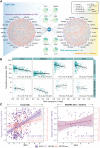

References
-
- Motzer R.J., Tannir N.M., McDermott D.F., Arén Frontera O., Melichar B., Choueiri T.K., Plimack E.R., Barthélémy P., Porta C., George S., et al. Nivolumab plus Ipilimumab versus Sunitinib in Advanced Renal-Cell Carcinoma. N. Engl. J. Med. 2018;378:1277–1290. doi: 10.1056/NEJMoa1712126. - DOI - PMC - PubMed
-
- Motzer R.J., Robbins P.B., Powles T., Albiges L., Haanen J.B., Larkin J., Mu X.J., Ching K.A., Uemura M., Pal S.K., et al. Avelumab plus axitinib versus sunitinib in advanced renal cell carcinoma: biomarker analysis of the phase 3 JAVELIN Renal 101 trial. Nat. Med. 2020;26:1733–1741. doi: 10.1038/s41591-020-1044-8. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

