Anticancer benzoxaboroles block pre-mRNA processing by directly inhibiting CPSF3
- PMID: 37967558
- PMCID: PMC10841686
- DOI: 10.1016/j.chembiol.2023.10.019
Anticancer benzoxaboroles block pre-mRNA processing by directly inhibiting CPSF3
Abstract
A novel class of benzoxaboroles was reported to induce cancer cell death but the mechanism was unknown. Using a forward genetics platform, we discovered mutations in cleavage and polyadenylation specific factor 3 (CPSF3) that reduce benzoxaborole binding and confer resistance. CPSF3 is the endonuclease responsible for pre-mRNA 3'-end processing, which is also important for RNA polymerase II transcription termination. Benzoxaboroles inhibit this endonuclease activity of CPSF3 in vitro and also curb transcriptional termination in cells, which results in the downregulation of numerous constitutively expressed genes. Furthermore, we used X-ray crystallography to demonstrate that benzoxaboroles bind to the active site of CPSF3 in a manner distinct from the other known inhibitors of CPSF3. The benzoxaborole compound impeded the growth of cancer cell lines derived from different lineages. Our results suggest benzoxaboroles may represent a promising lead as CPSF3 inhibitors for clinical development.
Copyright © 2023 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



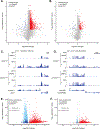
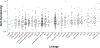
Similar articles
-
Clinical and veterinary trypanocidal benzoxaboroles target CPSF3.Proc Natl Acad Sci U S A. 2018 Sep 18;115(38):9616-9621. doi: 10.1073/pnas.1807915115. Epub 2018 Sep 5. Proc Natl Acad Sci U S A. 2018. PMID: 30185555 Free PMC article.
-
JTE-607, a multiple cytokine production inhibitor, targets CPSF3 and inhibits pre-mRNA processing.Biochem Biophys Res Commun. 2019 Oct 8;518(1):32-37. doi: 10.1016/j.bbrc.2019.08.004. Epub 2019 Aug 6. Biochem Biophys Res Commun. 2019. PMID: 31399191
-
A potent antimalarial benzoxaborole targets a Plasmodium falciparum cleavage and polyadenylation specificity factor homologue.Nat Commun. 2017 Mar 6;8:14574. doi: 10.1038/ncomms14574. Nat Commun. 2017. PMID: 28262680 Free PMC article.
-
On the Cutting Edge: Regulation and Therapeutic Potential of the mRNA 3' End Nuclease.Trends Biochem Sci. 2021 Sep;46(9):772-784. doi: 10.1016/j.tibs.2021.04.003. Epub 2021 Apr 30. Trends Biochem Sci. 2021. PMID: 33941430 Free PMC article. Review.
-
Recent molecular insights into canonical pre-mRNA 3'-end processing.Transcription. 2020 Apr;11(2):83-96. doi: 10.1080/21541264.2020.1777047. Epub 2020 Jun 11. Transcription. 2020. PMID: 32522085 Free PMC article. Review.
Cited by
-
Phenylboronic acid in targeted cancer therapy and diagnosis.Theranostics. 2025 Mar 3;15(9):3733-3748. doi: 10.7150/thno.104558. eCollection 2025. Theranostics. 2025. PMID: 40213663 Free PMC article. Review.
-
Small molecule inhibition of CPSF3 impacts R-loop distribution and abundance.bioRxiv [Preprint]. 2025 May 7:2025.05.07.652284. doi: 10.1101/2025.05.07.652284. bioRxiv. 2025. PMID: 40654756 Free PMC article. Preprint.
-
Aberrant pre-mRNA processing in cancer.J Exp Med. 2024 Nov 4;221(11):e20230891. doi: 10.1084/jem.20230891. Epub 2024 Sep 24. J Exp Med. 2024. PMID: 39316554 Free PMC article. Review.
References
-
- Nguyen TP, Fang M, Kim J, Wang B, Lin E, Khivansara V, Barrows N, Rivera-Cancel G, Goralski M, Cervantes CL, et al. (2023). Inducible mismatch repair streamlines forward genetic approaches to target identification of cytotoxic small molecules. Cell Chem Biol. 10.1016/j.chembiol.2023.07.017. - DOI - PMC - PubMed
-
- Akama T, Baker SJ, Zhang YK, Hernandez V, Zhou H, Sanders V, Freund Y, Kimura R, Maples KR, and Plattner JJ (2009). Discovery and structure-activity study of a novel benzoxaborole anti-inflammatory agent (AN2728) for the potential topical treatment of psoriasis and atopic dermatitis. Bioorganic & medicinal chemistry letters 19, 2129–2132. 10.1016/j.bmcl.2009.03.007. - DOI - PubMed
-
- Zhang YK, Plattner JJ, Freund YR, Easom EE, Zhou Y, Gut J, Rosenthal PJ, Waterson D, Gamo FJ, Angulo-Barturen I, et al. (2011). Synthesis and structure-activity relationships of novel benzoxaboroles as a new class of antimalarial agents. Bioorganic & medicinal chemistry letters 21, 644–651. 10.1016/j.bmcl.2010.12.034. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

