Mucosal-associated invariant T cells contribute to suppression of inflammatory myeloid cells in immune-mediated kidney disease
- PMID: 37968302
- PMCID: PMC10651937
- DOI: 10.1038/s41467-023-43269-0
Mucosal-associated invariant T cells contribute to suppression of inflammatory myeloid cells in immune-mediated kidney disease
Abstract
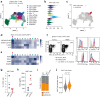
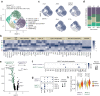
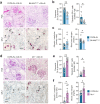
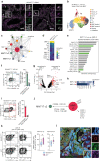
Mucosal-associated invariant T (MAIT) cells have been implicated in various inflammatory diseases of barrier organs, but so far, their role in kidney disease is unclear. Here we report that MAIT cells that recognize their prototypical ligand, the vitamin B2 intermediate 5-OP-RU presented by MR1, reside in human and mouse kidneys. Single cell RNAseq analysis reveals several intrarenal MAIT subsets, and one, carrying the genetic fingerprint of tissue-resident MAIT17 cells, is activated and expanded in a murine model of crescentic glomerulonephritis (cGN). An equivalent subset is also present in kidney biopsies of patients with anti-neutrophil cytoplasmatic antibody (ANCA)-associated cGN. MAIT cell-deficient MR1 mice show aggravated disease, whereas B6-MAITCAST mice, harboring higher MAIT cell numbers, are protected from cGN. The expanded MAIT17 cells express anti-inflammatory mediators known to suppress cGN, such as CTLA-4, PD-1, and TGF-β. Interactome analysis predicts CXCR6 - CXCL16-mediated cross-talk with renal mononuclear phagocytes, known to drive cGN progression. In line, we find that cGN is aggravated upon CXCL16 blockade. Finally, we present an optimized 5-OP-RU synthesis method which we apply to attenuating cGN in mice. In summary, we propose that CXCR6+ MAIT cells might play a protective role in cGN, implicating them as a potential target for anti-inflammatory therapies.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

