The extracellular matrix dictates regional competence for tumour initiation
- PMID: 37968399
- PMCID: PMC7615367
- DOI: 10.1038/s41586-023-06740-y
The extracellular matrix dictates regional competence for tumour initiation
Abstract
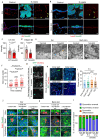
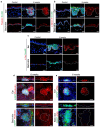
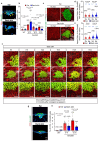
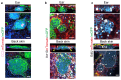
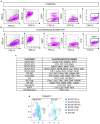
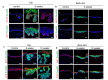
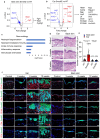
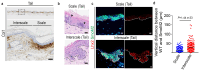
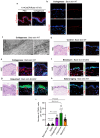
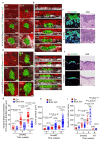
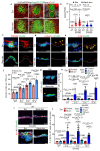
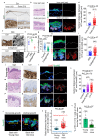
The skin epidermis is constantly renewed throughout life1,2. Disruption of the balance between renewal and differentiation can lead to uncontrolled growth and tumour initiation3. However, the ways in which oncogenic mutations affect the balance between renewal and differentiation and lead to clonal expansion, cell competition, tissue colonization and tumour development are unknown. Here, through multidisciplinary approaches that combine in vivo clonal analysis using intravital microscopy, single-cell analysis and functional analysis, we show how SmoM2-a constitutively active oncogenic mutant version of Smoothened (SMO) that induces the development of basal cell carcinoma-affects clonal competition and tumour initiation in real time. We found that expressing SmoM2 in the ear epidermis of mice induced clonal expansion together with tumour initiation and invasion. By contrast, expressing SmoM2 in the back-skin epidermis led to a clonal expansion that induced lateral cell competition without dermal invasion and tumour formation. Single-cell analysis showed that oncogene expression was associated with a cellular reprogramming of adult interfollicular cells into an embryonic hair follicle progenitor (EHFP) state in the ear but not in the back skin. Comparisons between the ear and the back skin revealed that the dermis has a very different composition in these two skin types, with increased stiffness and a denser collagen I network in the back skin. Decreasing the expression of collagen I in the back skin through treatment with collagenase, chronic UV exposure or natural ageing overcame the natural resistance of back-skin basal cells to undergoing EHFP reprogramming and tumour initiation after SmoM2 expression. Altogether, our study shows that the composition of the extracellular matrix regulates how susceptible different regions of the body are to tumour initiation and invasion.
© 2023. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
No competing interests.
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

