Elucidation of the genetic determination of clutch traits in Chinese local chickens of the Laiwu Black breed
- PMID: 37968610
- PMCID: PMC10652520
- DOI: 10.1186/s12864-023-09798-0
Elucidation of the genetic determination of clutch traits in Chinese local chickens of the Laiwu Black breed
Abstract
Background: Egg laying rate (LR) is associated with a clutch, which is defined as consecutive days of oviposition. The clutch trait can be used as a selection indicator to improve egg production in poultry breeding. However, little is known about the genetic basis of clutch traits. In this study, our aim was to estimate genetic parameters and identify quantitative trait single nucleotide polymorphisms for clutch traits in 399 purebred Laiwu Black chickens (a native Chinese breed) using a genome-wide association study (GWAS).
Methods: In this work, after estimating the genetic parameters of age at first egg, body weight at first egg, LR, longest clutch until 52 week of age, first week when the longest clutch starts, last week when the longest clutch ends, number of clutches, and longest number of days without egg-laying until 52 week of age, we identified single nucleotide polymorphisms (SNPs) and potential candidate genes associated with clutch traits in Laiwu Black chickens. The restricted maximum likelihood method was used to estimate genetic parameters of clutch pattern in 399 Laiwu Black hens, using the GCTA software.
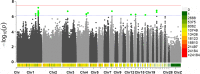
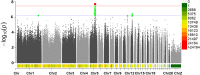
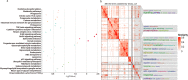
Results: The results showed that SNP-based heritability estimates of clutch traits ranged from 0.06 to 0.59. Genotyping data were obtained from whole genome re-sequencing data. After quality control, a total of 10,810,544 SNPs remained to be analyzed. The GWAS revealed that 421 significant SNPs responsible for clutch traits were scattered on chicken chromosomes 1-14, 17-19, 21-25, 28 and Z. Among the annotated genes, NELL2, SMYD9, SPTLC2, SMYD3 and PLCL1 were the most promising candidates for clutch traits in Laiwu Black chickens.
Conclusion: The findings of this research provide critical insight into the genetic basis of clutch traits. These results contribute to the identification of candidate genes and variants. Genes and SNPs potentially provide new avenues for further research and would help to establish a framework for new methods of genomic prediction, and increase the accuracy of estimated genetic merit for egg production and clutch traits.
Keywords: Chicken; Clutch traits; GWAS; Genome; Laying rate.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
Genome-wide association studies of egg production traits by whole genome sequencing of Laiwu Black chicken.Poult Sci. 2024 Jun;103(6):103705. doi: 10.1016/j.psj.2024.103705. Epub 2024 Mar 31. Poult Sci. 2024. PMID: 38598913 Free PMC article.
-
Comparison of genomic prediction accuracy using different models for egg production traits in Taiwan country chicken.Poult Sci. 2024 Oct;103(10):104063. doi: 10.1016/j.psj.2024.104063. Epub 2024 Jul 6. Poult Sci. 2024. PMID: 39098301 Free PMC article.
-
Novel Polymorphisms in RAPGEF6 Gene Associated with Egg-Laying Rate in Chinese Jing Hong Chicken using Genome-Wide SNP Scan.Genes (Basel). 2019 May 20;10(5):384. doi: 10.3390/genes10050384. Genes (Basel). 2019. PMID: 31137587 Free PMC article.
-
Endocrine and genetic factors affecting egg laying performance in chickens: a review.Br Poult Sci. 2020 Oct;61(5):538-549. doi: 10.1080/00071668.2020.1758299. Epub 2020 Jun 1. Br Poult Sci. 2020. PMID: 32306752 Review.
-
The study of candidate genes in the improvement of egg production in ducks - a review.Poult Sci. 2022 Jul;101(7):101850. doi: 10.1016/j.psj.2022.101850. Epub 2022 Mar 11. Poult Sci. 2022. PMID: 35544958 Free PMC article. Review.
Cited by
-
Genome-wide analyses reveal intricate genetic mechanisms underlying egg production efficiency in chickens.J Anim Sci Biotechnol. 2025 Aug 11;16(1):114. doi: 10.1186/s40104-025-01245-2. J Anim Sci Biotechnol. 2025. PMID: 40790246 Free PMC article.
-
Integrating genomics and transcriptomics reveals candidate genes affecting loin muscle area in Huaxi cattle.PLoS One. 2025 May 9;20(5):e0322026. doi: 10.1371/journal.pone.0322026. eCollection 2025. PLoS One. 2025. PMID: 40344088 Free PMC article.
-
Synergistic transcriptomic and metabolomic analyses in Zi geese ovaries with different clutch lengths.Poult Sci. 2025 Jul;104(7):105210. doi: 10.1016/j.psj.2025.105210. Epub 2025 Apr 23. Poult Sci. 2025. PMID: 40294555 Free PMC article.
-
Genome-wide association analysis identified the involvement of MRPS22 in the regulation of Muscovy duck broodiness.Poult Sci. 2025 Apr;104(4):104994. doi: 10.1016/j.psj.2025.104994. Epub 2025 Mar 6. Poult Sci. 2025. PMID: 40068571 Free PMC article.
-
Genome-wide association studies of egg production traits by whole genome sequencing of Laiwu Black chicken.Poult Sci. 2024 Jun;103(6):103705. doi: 10.1016/j.psj.2024.103705. Epub 2024 Mar 31. Poult Sci. 2024. PMID: 38598913 Free PMC article.
References
-
- Lillpers K, Wilhelmson M. Genetic and phenotypic parameters for oviposition pattern traits in three selection lines of laying hens. Brit poultry sci. 1993;34(2):297–308. doi: 10.1080/00071669308417586. - DOI
-
- England A , Ruhnke I .The influence of light of different wavelengths on laying hen production and egg quality. World’s Poult Sci J. 2020:1–16. 10.1080/00439339.2020.1789023.
-
- Wolc A, Bednarczyk M, Lisowski M, Szwaczkowski T. Genetic relationships among time of egg formation, clutch traits and traditional selection traits in laying hens. J Anim Feed Sci. 2010;19(3):452–459. doi: 10.22358/jafs/66309/2010. - DOI
-
- Bednarczyk M, Kiecłzewski K, Szwaczkowski T. Genetic parameters of the traditional selection traits and some clutch traits in a commercial line of laying hens. Arch Geflügelk. 2000;64(3):129–133.
MeSH terms
Grants and funding
- 2019LZGC019/Agricultural Stock Breeding Project of Shandong Province
- CXGC2022C04/Agricultural Scientific and Technological Innovation Project of Shandong Academy of Agricultural Sciences
- CXGC2022D04/Agricultural Scientific and Technological Innovation Project of Shandong Academy of Agricultural Sciences
- 2020LZGC013/Agricultural Breed Project of Shandong Province
- ZR2020MC169/Shandong Provincial Natural Science Foundation
- CARS-41/China Agriculture Research System of MOF and MARA
- CARS-40/China Agriculture Research System of MOF and MARA
- 2021CXGC0010805/The Key Research Program of Shandong province
- CXGC2022E11/Agricultural Scientific and Technological Innovation Project of Shandong Academy of Agricultural Sciences
- CXGC2023F11/Agricultural Scientific and Technological Innovation Project of Shandong Academy of Agricultural Sciences
LinkOut - more resources
Full Text Sources

