Exploring the key genomic variation in monkeypox virus during the 2022 outbreak
- PMID: 37968621
- PMCID: PMC10652487
- DOI: 10.1186/s12863-023-01171-0
Exploring the key genomic variation in monkeypox virus during the 2022 outbreak
Abstract
Background: In 2022, a global outbreak of monkeypox occurred with a significant shift in its epidemiological characteristics. The monkeypox virus (MPXV) belongs to the B.1 lineage, and its genomic variations that were linked to the outbreak were investigated in this study. Previous studies have suggested that viral genomic variation plays a crucial role in the pathogenicity and transmissibility of viruses. Therefore, understanding the genomic variation of MPXV is crucial for controlling future outbreaks.
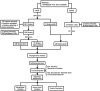
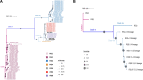
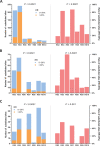
Methods: This study employed bioinformatics and phylogenetic approaches to evaluate the key genomic variation in the B.1 lineage of MPXV. A total of 979 MPXV strains were screened, and 212 representative strains were analyzed to identify specific substitutions in the viral genome. Reference sequences were constructed for each of the 10 lineages based on the most common nucleotide at each site. A total of 49 substitutions were identified, with 23 non-synonymous substitutions. Class I variants, which had significant effects on protein conformation likely to affect viral characteristics, were classified among the non-synonymous substitutions.
Results: The phylogenetic analysis revealed 10 relatively monophyletic branches. The study identified 49 substitutions specific to the B.1 lineage, with 23 non-synonymous substitutions that were classified into Class I, II, and III variants. The Class I variants were likely responsible for the observed changes in the characteristics of circulating MPXV in 2022. These key mutations, particularly Class I variants, played a crucial role in the pathogenicity and transmissibility of MPXV.
Conclusion: This study provides an understanding of the genomic variation of MPXV in the B.1 lineage linked to the recent outbreak of monkeypox. The identification of key mutations, particularly Class I variants, sheds light on the molecular mechanisms underlying the observed changes in the characteristics of circulating MPXV. Further studies can focus on functional domains affected by these mutations, enabling the development of effective control strategies against future monkeypox outbreaks.
Keywords: Genomic variation; Monkeypox virus; Phylogenetic tree; Protein structure prediction; Reference sequence.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

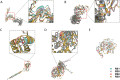
References
-
- Ullah A, Shahid FA, Haq MU, Tahir Ul Qamar M, Irfan M, Shaker B, Ahmad S, Alrumaihi F, Allemailem KS, Almatroudi A. An integrative reverse vaccinology, immunoinformatic, docking and simulation approaches towards designing of multi-epitopes based vaccine against monkeypox virus. J Biomol Struct Dyn. 2023;41:821-7834. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
