Invasive FoxM1 phosphorylated by PLK1 induces the polarization of tumor-associated macrophages to promote immune escape and metastasis, amplified by IFITM1
- PMID: 37968723
- PMCID: PMC10652615
- DOI: 10.1186/s13046-023-02872-1
Invasive FoxM1 phosphorylated by PLK1 induces the polarization of tumor-associated macrophages to promote immune escape and metastasis, amplified by IFITM1
Abstract
Background: Understanding the mechanism behind immune cell plasticity in cancer metastasis is crucial for identifying key regulators. Previously we found that mitotic factors regulate epithelial-mesenchymal transition, but how these factors convert to metastatic players in the tumor microenvironment (TME) is not fully understood.
Methods: The clinical importance of mitotic factors was analyzed by heatmap analysis, a KM plot, and immunohistochemistry in lung adenocarcinoma (LUAD) patients. Immunoprecipitation, LC-MS/MS, kinase assay, and site-directed mutagenesis were performed for the interaction and phosphorylation. A tail-vein injection mouse model, Transwell-based 3D culture, microarray analysis, coculture with monocytes, and chromatin immunoprecipitation assays were used to elucidate the function of phosphorylated FoxM1 in metastasis of TME.
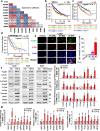
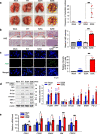
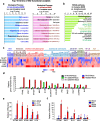
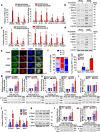
Results: The phosphorylated FoxM1 at Ser25 by PLK1 acquires the reprogramming ability to stimulate the invasive traits in cancer and influence immune cell plasticity. This invasive form of p-FoxM1 upregulates the expression of IL1A/1B, VEGFA, and IL6 by direct activation, recruiting monocytes and promoting the polarization of M2d-like tumor-associated macrophages (TAMs). Upregulation of PD-L1 in LUAD having phosphomimetic FoxM1 facilitates immune evasion. In invasive LUAD with phosphomimetic FoxM1, IFITM1 is the most highly expressed through the activation of the STING-TBK1-IRF3 signaling, which enhances FoxM1-mediated signaling. Clinically, higher expression of FOXM1, PLK1, and IFITM1 is inversely correlated with the survival rate of advanced LUAD patients, providing a promising therapeutic strategy for the treatment of LUAD.
Conclusion: FoxM1-based therapy would be a potential therapeutic strategy for LUAD to reduce TAM polarization, immune escape, and metastasis, since FoxM1 functions as a genetic reprogramming factor reinforcing LUAD malignancy in the TME.
Keywords: FoxM1; Invasiveness; PLK1; Phosphorylation; Tumor-associated macrophages.
© 2023. The Author(s).
Conflict of interest statement
The authors have declared that no conflict of interest exists.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

