Biocatalytic reductive amination as a route to isotopically labelled amino acids suitable for analysis of large proteins by NMR
- PMID: 37969586
- PMCID: PMC10631221
- DOI: 10.1039/d3sc01718d
Biocatalytic reductive amination as a route to isotopically labelled amino acids suitable for analysis of large proteins by NMR
Abstract
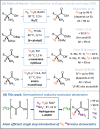
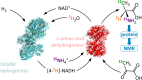
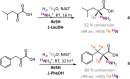
We demonstrate an atom-efficient and easy to use H2-driven biocatalytic platform for the enantioselective incorporation of 2H-atoms into amino acids. By combining the biocatalytic deuteration catalyst with amino acid dehydrogenase enzymes capable of reductive amination, we synthesised a library of multiply isotopically labelled amino acids from low-cost isotopic precursors, such as 2H2O and 15NH4+. The chosen approach avoids the use of pre-labeled 2H-reducing agents, and therefore vastly simplifies product cleanup. Notably, this strategy enables 2H, 15N, and an asymmetric centre to be introduced at a molecular site in a single step, with full selectivity, under benign conditions, and with near 100% atom economy. The method facilitates the preparation of amino acid isotopologues on a half-gram scale. These amino acids have wide applicability in the analytical life sciences, and in particular for NMR spectroscopic analysis of proteins. To demonstrate the benefits of the approach for enabling the workflow of protein NMR chemists, we prepared l-[α-2H,15N, β-13C]-alanine and integrated it into a large (>400 kDa) heat-shock protein oligomer, which was subsequently analysable by methyl-TROSY techniques, revealing new structural information.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
A patent application by J. S. R., H. A. R. and K. A. V. detailing some of this research was filed through Oxford University Innovation (Feb 2018). The remaining authors declare no competing interests.
Figures



References
-
- Kelman Z., Isotope Labeling of Biomolecules – Applications, Elsevier, New York, 2016, vol. 566
-
- Kainosho M., Miyanoiri Y. and Takeda M., in Experimental Approaches of NMR Spectroscopy, Springer Singapore, Singapore, 2018, pp. 37–61
-
- Ashkar R. Bilheux H. Z. Bordallo H. Briber R. Callaway D. J. E. Cheng X. Chu X.-Q. Curtis J. E. Dadmun M. Fenimore P. Fushman D. Gabel F. Gupta K. Herberle F. Heinrich F. Hong L. Katsaras J. Kelman Z. Kharlampieva E. Kneller G. R. Kovalevsky A. Krueger S. Langan P. Lieberman R. Liu Y. Losche M. Lyman E. Mao Y. Marino J. Mattos C. Meilleur F. Moody P. Nickels J. D. O'Dell W. B. O'Neill H. Perez-Salas U. Peters J. Petridis L. Sokolov A. P. Stanley C. Wagner N. Weinrich M. Weiss K. Wymore T. Zhang Y. Smith J. C. Acta Crystallogr., Sect. D: Struct. Biol. 2018;74:1129–1168. doi: 10.1107/S2059798318017503. - DOI - PubMed
LinkOut - more resources
Full Text Sources

