A single-cell atlas depicting the cellular and molecular features in human anterior cruciate ligamental degeneration: A single cell combined spatial transcriptomics study
- PMID: 37970848
- PMCID: PMC10688969
- DOI: 10.7554/eLife.85700
A single-cell atlas depicting the cellular and molecular features in human anterior cruciate ligamental degeneration: A single cell combined spatial transcriptomics study
Abstract
Background: To systematically identify cell types in the human ligament, investigate how ligamental cell identities, functions, and interactions participated in the process of ligamental degeneration, and explore the changes of ligamental microenvironment homeostasis in the disease progression.
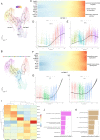
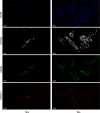
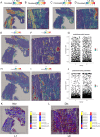
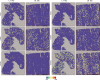
Methods: Using single-cell RNA sequencing and spatial RNA sequencing of approximately 49,356 cells, we created a comprehensive cell atlas of healthy and degenerated human anterior cruciate ligaments. We explored the variations of the cell subtypes' spatial distributions and the different processes involved in the disease progression, linked them with the ligamental degeneration process using computational analysis, and verified findings with immunohistochemical and immunofluorescent staining.
Results: We identified new fibroblast subgroups that contributed to the disease, mapped out their spatial distribution in the tissue and revealed two dynamic trajectories in the process of the degenerative process. We compared the cellular interactions between different tissue states and identified important signaling pathways that may contribute to the disease.
Conclusions: This cell atlas provides the molecular foundation for investigating how ligamental cell identities, biochemical functions, and interactions contributed to the ligamental degeneration process. The discoveries revealed the pathogenesis of ligamental degeneration at the single-cell and spatial level, which is characterized by extracellular matrix remodeling. Our results provide new insights into the control of ligamental degeneration and potential clues to developing novel diagnostic and therapeutic strategies.
Funding: This study was funded by the National Natural Science Foundation of China (81972123, 82172508, 82372490) and 1.3.5 Project for Disciplines of Excellence of West China Hospital Sichuan University (ZYJC21030, ZY2017301).
Keywords: extracellular matrix remodeling; human; ligamental degeneration; medicine; single-cell RNA-seq; spatial RNA-seq.
© 2023, Yang et al.
Conflict of interest statement
RY, TX, LZ, MG, LY, JL, WF No competing interests declared
Figures







Update of
- doi: 10.1101/2023.01.05.23284219
References
-
- Busch C, Girke G, Kohl B, Stoll C, Lemke M, Krasnici S, Ertel W, Silawal S, John T, Schulze-Tanzil G. Complement gene expression is regulated by pro-inflammatory cytokines and the anaphylatoxin C3a in human tenocytes. Molecular Immunology. 2013;53:363–373. doi: 10.1016/j.molimm.2012.09.001. - DOI - PubMed
-
- Ferrão PM, Nisimura LM, Moreira OC, Land MG, Pereira MC, Mendonça-Lima L, Araujo-Jorge TC, Waghabi MC, Garzoni LR. Inhibition of TGF-β pathway reverts extracellular matrix remodeling in T. cruzi-infected cardiac spheroids. Experimental Cell Research. 2018;362:260–267. doi: 10.1016/j.yexcr.2017.11.026. - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

