Heparin-binding epidermal growth factor and fibroblast growth factor 2 rescue Müller glia-derived progenitor cell formation in microglia- and macrophage-ablated chick retinas
- PMID: 37971210
- PMCID: PMC10730090
- DOI: 10.1242/dev.202070
Heparin-binding epidermal growth factor and fibroblast growth factor 2 rescue Müller glia-derived progenitor cell formation in microglia- and macrophage-ablated chick retinas
Abstract
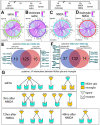
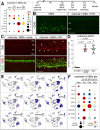
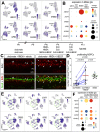
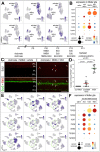
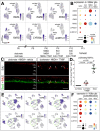
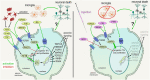
Recent studies have demonstrated the impact of pro-inflammatory signaling and reactive microglia/macrophages on the formation of Müller glial-derived progenitor cells (MGPCs) in the retina. In chick retina, ablation of microglia/macrophages prevents the formation of MGPCs. Analyses of single-cell RNA-sequencing chick retinal libraries revealed that quiescent and activated microglia/macrophages have a significant impact upon the transcriptomic profile of Müller glia (MG). In damaged monocyte-depleted retinas, MG fail to upregulate genes related to different cell signaling pathways, including those related to Wnt, heparin-binding epidermal growth factor (HBEGF), fibroblast growth factor (FGF) and retinoic acid receptors. Inhibition of GSK3β, to simulate Wnt signaling, failed to rescue the deficit in MGPC formation, whereas application of HBEGF or FGF2 completely rescued the formation of MGPCs in monocyte-depleted retinas. Inhibition of Smad3 or activation of retinoic acid receptors partially rescued the formation of MGPCs in monocyte-depleted retinas. We conclude that signals produced by reactive microglia/macrophages in damaged retinas stimulate MG to upregulate cell signaling through HBEGF, FGF and retinoic acid, and downregulate signaling through TGFβ/Smad3 to promote the reprogramming of MG into proliferating MGPCs.
Keywords: Cell signaling; Microglia; Müller glia; Retinal regeneration; scRNA-seq.
© 2023. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures


References
-
- Campbell, W. A., Deshmukh, A., Blum, S., Todd, L., Mendonca, N., Weist, J., Zent, J., Hoang, T. V., Blackshaw, S., Leight, J.et al. (2019). Matrix-metalloproteinase expression and gelatinase activity in the avian retina and their influence on Müller glia proliferation. Exp. Neurol. 320, 112984. 10.1016/j.expneurol.2019.112984 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

