With a pinch of salt: metagenomic insights into Namib Desert salt pan microbial mats and halites reveal functionally adapted and competitive communities
- PMID: 37971255
- PMCID: PMC10734447
- DOI: 10.1128/aem.00629-23
With a pinch of salt: metagenomic insights into Namib Desert salt pan microbial mats and halites reveal functionally adapted and competitive communities
Abstract
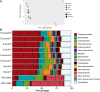
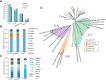
The hyperarid Namib Desert is one of the oldest deserts on Earth. It contains multiple clusters of playas which are saline-rich springs surrounded by halite evaporites. Playas are of great ecological importance, and their indigenous (poly)extremophilic microorganisms are potentially involved in the precipitation of minerals such as carbonates and sulfates and have been of great biotechnological importance. While there has been a considerable amount of microbial ecology research performed on various Namib Desert edaphic microbiomes, little is known about the microbial communities inhabiting its multiple playas. In this work, we provide a comprehensive taxonomic and functional potential characterization of the microbial, including viral, communities of sediment mats and halites from two distant salt pans of the Namib Desert, contributing toward a better understanding of the ecology of this biome.
Keywords: CRISPR-Cas; functional diversity; gene transfer agent; halite; horizontal gene transfer; microbial mat; playa; salt pan; taxonomic diversity; virus-host interactions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Soil fungal diversity and assembly along a xeric stress gradient in the central Namib Desert.Fungal Biol. 2023 Apr;127(4):997-1003. doi: 10.1016/j.funbio.2023.03.001. Epub 2023 Mar 7. Fungal Biol. 2023. PMID: 37024159
-
Namib Desert edaphic bacterial, fungal and archaeal communities assemble through deterministic processes but are influenced by different abiotic parameters.Extremophiles. 2017 Mar;21(2):381-392. doi: 10.1007/s00792-016-0911-1. Epub 2017 Jan 5. Extremophiles. 2017. PMID: 28058513
-
Namib Desert Soil Microbial Community Diversity, Assembly, and Function Along a Natural Xeric Gradient.Microb Ecol. 2018 Jan;75(1):193-203. doi: 10.1007/s00248-017-1009-8. Epub 2017 Jun 24. Microb Ecol. 2018. PMID: 28647755
-
Microbiomics of Namib Desert habitats.Extremophiles. 2020 Jan;24(1):17-29. doi: 10.1007/s00792-019-01122-7. Epub 2019 Aug 2. Extremophiles. 2020. PMID: 31376000 Review.
-
Diversity and Ecology of Viruses in Hyperarid Desert Soils.Appl Environ Microbiol. 2015 Nov 20;82(3):770-7. doi: 10.1128/AEM.02651-15. Print 2016 Feb 1. Appl Environ Microbiol. 2015. PMID: 26590289 Free PMC article. Review.
Cited by
-
Functional redundancy buffers the effect of poly-extreme environmental conditions on southern African dryland soil microbial communities.FEMS Microbiol Ecol. 2024 Nov 23;100(12):fiae157. doi: 10.1093/femsec/fiae157. FEMS Microbiol Ecol. 2024. PMID: 39568064 Free PMC article.
-
South Africa's indigenous microbial diversity for industrial applications: A review of the current status and opportunities.Heliyon. 2023 Jun 1;9(6):e16723. doi: 10.1016/j.heliyon.2023.e16723. eCollection 2023 Jun. Heliyon. 2023. PMID: 37484259 Free PMC article. Review.
-
Diversity of Microbial Mats in the Makgadikgadi Salt Pans, Botswana.Microorganisms. 2024 Jan 11;12(1):147. doi: 10.3390/microorganisms12010147. Microorganisms. 2024. PMID: 38257974 Free PMC article.
References
-
- Bryant RG. 1996. Validated linear mixture modelling of landsat TM data for mapping evaporite minerals on a playa surface: methods and applications. Int J Remote Sens 17:315–330. doi:10.1080/01431169608949008 - DOI
-
- Waiser MJ, Robarts RD.. 2009. Saline inland waters, p. 634–644. In Likens, GE (ed.), Encyclopedia of inland waters. Academic Press, Oxford.
-
- Öztürk M, Böer B, Barth H-J, Clüsener-Godt M, Khan MA, Breckle S-W. 2011. Sabkha Ecosystems. In Sabkha Ecosystems. Dordrecht. doi:10.1007/978-90-481-9673-9 - DOI
-
- Ward JD, Seely MK, Lancaster N. 1983. On the antiquity of the namib south African. J Sci 79:9.
-
- Day JA, Seely MK. 1988. Physical and chemical conditions in an hypersaline spring in the namib desert. Hydrobio 160:141–153. doi:10.1007/BF00015477 - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

