Cytomegalovirus-vectored COVID-19 vaccines elicit neutralizing antibodies against the SARS-CoV-2 Omicron variant (BA.2) in mice
- PMID: 37971259
- PMCID: PMC10883801
- DOI: 10.1128/spectrum.02463-23
Cytomegalovirus-vectored COVID-19 vaccines elicit neutralizing antibodies against the SARS-CoV-2 Omicron variant (BA.2) in mice
Abstract
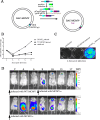
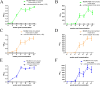
Cytomegalovirus (CMV) has been used as a novel viral vector for vaccine development and gene therapy. Coronavirus disease 2019 is an infectious disease caused by the SARS-CoV-2 virus, which is highly mutable and is still circulating globally. The study showed that the CMV viral vector caused transient systemic infection and induced robust transgene expression in vivo. CMV vectors expressing different SARS-CoV-2 proteins were immunogenic and could elicit neutralizing antibodies against a highly mutated Omicron variant (BA.2). The expression level of receptor-binding domain (RBD) protein was higher than that of full-length S protein using CMV as a vaccine vector, and CMV vector expression RBD protein elicited higher RBD-binding and neutralizing antibodies. Moreover, the study showed that CMV-vectored vaccines would not cause unexpected viral transmission, and pre-existing immunity might impair the immunogenicity of subsequent CMV-vectored vaccines. These works provide meaningful insights for the development of a CMV-based vector vaccine platform and the prevention and control strategies for SARS-CoV-2 infection.
Keywords: SARS-CoV-2; cytomegalovirus; immunogenicity evaluation; variant of concern; viral vector-based vaccine.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Wu F, Zhao S, Yu B, Chen Y-M, Wang W, Song Z-G, Hu Y, Tao Z-W, Tian J-H, Pei Y-Y, Yuan M-L, Zhang Y-L, Dai F-H, Liu Y, Wang Q-M, Zheng J-J, Xu L, Holmes EC, Zhang Y-Z. 2020. A new coronavirus associated with human respiratory disease in China. Nature 580:265–269. doi: 10.1038/s41586-020-2202-3 - DOI - PMC - PubMed
-
- Wu S, Zhong G, Zhang J, Shuai L, Zhang Z, Wen Z, Wang B, Zhao Z, Song X, Chen Y, Liu R, Fu L, Zhang J, Guo Q, Wang C, Yang Y, Fang T, Lv P, Wang J, Xu J, Li J, Yu C, Hou L, Bu Z, Chen W. 2020. A single dose of an adenovirus-vectored vaccine provides protection against SARS-CoV-2 challenge. Nat Commun 11:4081. doi: 10.1038/s41467-020-17972-1 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

