Structural and biochemical insights into NEIL2's preference for abasic sites
- PMID: 37971311
- PMCID: PMC10711445
- DOI: 10.1093/nar/gkad1075
Structural and biochemical insights into NEIL2's preference for abasic sites
Abstract
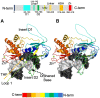
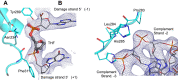
Cellular DNA is subject to damage from a multitude of sources and repair or bypass of sites of damage utilize an array of context or cell cycle dependent systems. The recognition and removal of oxidatively damaged bases is the task of DNA glycosylases from the base excision repair pathway utilizing two structural families that excise base lesions in a wide range of DNA contexts including duplex, single-stranded and bubble structures arising during transcription. The mammalian NEIL2 glycosylase of the Fpg/Nei family excises lesions from each of these DNA contexts favoring the latter two with a preference for oxidized cytosine products and abasic sites. We have determined the first liganded crystal structure of mammalian NEIL2 in complex with an abasic site analog containing DNA duplex at 2.08 Å resolution. Comparison to the unliganded structure revealed a large interdomain conformational shift upon binding the DNA substrate accompanied by local conformational changes in the C-terminal domain zinc finger and N-terminal domain void-filling loop necessary to position the enzyme on the DNA. The detailed biochemical analysis of NEIL2 with an array of oxidized base lesions indicates a significant preference for its lyase activity likely to be paramount when interpreting the biological consequences of variants.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures







Similar articles
-
Requirements for DNA bubble structure for efficient cleavage by helix-two-turn-helix DNA glycosylases.Mutagenesis. 2020 Feb 13;35(1):119-128. doi: 10.1093/mutage/gez047. Mutagenesis. 2020. PMID: 31784740
-
Structural investigation of a viral ortholog of human NEIL2/3 DNA glycosylases.DNA Repair (Amst). 2013 Dec;12(12):1062-71. doi: 10.1016/j.dnarep.2013.09.004. Epub 2013 Oct 10. DNA Repair (Amst). 2013. PMID: 24120312 Free PMC article.
-
Unique Structural Features of Mammalian NEIL2 DNA Glycosylase Prime Its Activity for Diverse DNA Substrates and Environments.Structure. 2021 Jan 7;29(1):29-42.e4. doi: 10.1016/j.str.2020.08.001. Epub 2020 Aug 25. Structure. 2021. PMID: 32846144 Free PMC article.
-
Functional roles and cancer variants of the bifunctional glycosylase NEIL2.Environ Mol Mutagen. 2024 Apr;65 Suppl 1:40-56. doi: 10.1002/em.22555. Epub 2023 Jun 26. Environ Mol Mutagen. 2024. PMID: 37310399 Review.
-
DNA glycosylases search for and remove oxidized DNA bases.Environ Mol Mutagen. 2013 Dec;54(9):691-704. doi: 10.1002/em.21820. Epub 2013 Oct 7. Environ Mol Mutagen. 2013. PMID: 24123395 Free PMC article. Review.
Cited by
-
Research progress on the role of the NEIL family in cancer.Front Cell Dev Biol. 2025 Jul 21;13:1612329. doi: 10.3389/fcell.2025.1612329. eCollection 2025. Front Cell Dev Biol. 2025. PMID: 40761744 Free PMC article. Review.
-
NEIL3: A unique DNA glycosylase involved in interstrand DNA crosslink repair.DNA Repair (Amst). 2024 Jul;139:103680. doi: 10.1016/j.dnarep.2024.103680. Epub 2024 Apr 21. DNA Repair (Amst). 2024. PMID: 38663144 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

