A proxitome-RNA-capture approach reveals that processing bodies repress coregulated hub genes
- PMID: 37971938
- PMCID: PMC10896293
- DOI: 10.1093/plcell/koad288
A proxitome-RNA-capture approach reveals that processing bodies repress coregulated hub genes
Abstract
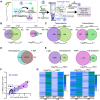
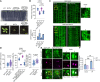
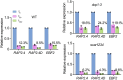
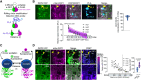
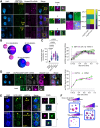
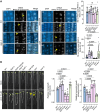
Cellular condensates are usually ribonucleoprotein assemblies with liquid- or solid-like properties. Because these subcellular structures lack a delineating membrane, determining their compositions is difficult. Here we describe a proximity-biotinylation approach for capturing the RNAs of the condensates known as processing bodies (PBs) in Arabidopsis (Arabidopsis thaliana). By combining this approach with RNA detection, in silico, and high-resolution imaging approaches, we studied PBs under normal conditions and heat stress. PBs showed a much more dynamic RNA composition than the total transcriptome. RNAs involved in cell wall development and regeneration, plant hormonal signaling, secondary metabolism/defense, and RNA metabolism were enriched in PBs. RNA-binding proteins and the liquidity of PBs modulated RNA recruitment, while RNAs were frequently recruited together with their encoded proteins. In PBs, RNAs follow distinct fates: in small liquid-like PBs, RNAs get degraded while in more solid-like larger ones, they are stored. PB properties can be regulated by the actin-polymerizing SCAR (suppressor of the cyclic AMP)-WAVE (WASP family verprolin homologous) complex. SCAR/WAVE modulates the shuttling of RNAs between PBs and the translational machinery, thereby adjusting ethylene signaling. In summary, we provide an approach to identify RNAs in condensates that allowed us to reveal a mechanism for regulating RNA fate.
© The Author(s) 2023. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Conflict of interest statement
Conflict of interest statement. None declared.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

