Exploration of the B3 transcription factor superfamily in Aquilaria sinensis reveal their involvement in seed recalcitrance and agarwood formation
- PMID: 37972007
- PMCID: PMC10653465
- DOI: 10.1371/journal.pone.0294358
Exploration of the B3 transcription factor superfamily in Aquilaria sinensis reveal their involvement in seed recalcitrance and agarwood formation
Abstract
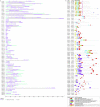
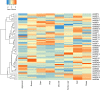
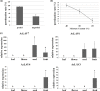
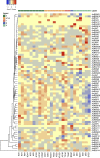
The endangered tree species of the Aquilaria genus produce agarwood, a high value material produced only after wounding; however, conservation of Aquilaria seeds is difficult. The B3 transcription factor family has diverse important functions in plant development, especially in seed development, although their functions in other areas, such as stress responses, remain to be revealed. Here germination tests proved that the seeds of A. sinensis were recalcitrant seeds. To provide insights into the B3 superfamily, the members were identified and characterized by bioinformatic approaches and classified by phylogenetic analysis and domain structure. In total, 71 members were identified and classified into four subfamilies. Each subfamily not only had similar domains, but also had conserved motifs in their B3 domains. For the seed-related LAV subfamily, the B3 domain of AsLAV3 was identical to that of AsVALs but lacked a typical zf-CW domain such as VALs. AsLAV5 lacks a typical PHD-L domain present in Arabidopsis VALs. qRT-PCR expression analysis showed that the LEC2 ortholog AsLAV4 was not expressed in seeds. RAVs and REMs induced after wound treatment were also identified. These findings provide insights into the functions of B3 genes and seed recalcitrance of A. sinensis and indicate the role of B3 genes in wound response and agarwood formation.This is the first work to investigate the B3 family in A. sinensis and to provide insights of the molecular mechanism of seed recalcitrance.This will be a valuable guidance for studies of B3 genes in stress responses, secondary metabolite biosynthesis, and seed development.
Copyright: © 2023 Jin et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- López-Sampson A, Page T. History of use and trade of agarwood. Econ Bot. 2018;72(1):107–29.
-
- The International Union for Conservation of Nature. IUCN Red List of Threatened Species. In.;2008.
-
- the Convention on International Trade in Endangered Species of Wild Fauna and Flora. “Amendments to appendices I and II of CITES" In Proceedings of Thirteenth Meeting of the Conference of the Parties. Johannesburg, South Africa. 2017.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

