Tunable force transduction through the Escherichia coli cell envelope
- PMID: 37972066
- PMCID: PMC10666116
- DOI: 10.1073/pnas.2306707120
Tunable force transduction through the Escherichia coli cell envelope
Abstract
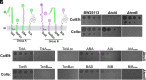
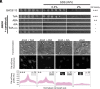
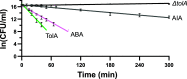
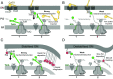
The outer membrane (OM) of Gram-negative bacteria is not energised and so processes requiring a driving force must connect to energy-transduction systems in the inner membrane (IM). Tol (Tol-Pal) and Ton are related, proton motive force- (PMF-) coupled assemblies that stabilise the OM and import essential nutrients, respectively. Both rely on proton-harvesting IM motor (stator) complexes, which are homologues of the flagellar stator unit Mot, to transduce force to the OM through elongated IM force transducer proteins, TolA and TonB, respectively. How PMF-driven motors in the IM generate mechanical work at the OM via force transducers is unknown. Here, using cryoelectron microscopy, we report the 4.3Å structure of the Escherichia coli TolQR motor complex. The structure reaffirms the 5:2 stoichiometry seen in Ton and Mot and, with motor subunits related to each other by 10 to 16° rotation, supports rotary motion as the default for these complexes. We probed the mechanism of force transduction to the OM through in vivo assays of chimeric TolA/TonB proteins where sections of their structurally divergent, periplasm-spanning domains were swapped or replaced by an intrinsically disordered sequence. We find that TolA mutants exhibit a spectrum of force output, which is reflected in their respective abilities to both stabilise the OM and import cytotoxic colicins across the OM. Our studies demonstrate that structural rigidity of force transducer proteins, rather than any particular structural form, drives the efficient conversion of PMF-driven rotary motions of 5:2 motor complexes into physiologically relevant force at the OM.
Keywords: cell envelope; force transduction; gram-negative bacteria; outer membrane.
Conflict of interest statement
C.K. owns equity in antibiotics development company Glox Therapeutics Ltd. All other authors declares no competing interests.
Figures

References
-
- Lithgow T., Stubenrauch C. J., Stumpf M. P. H., Surveying membrane landscapes: A new look at the bacterial cell surface. Nat. Rev. Microbiol. 21, 502–518 (2023). - PubMed
-
- Lazzaroni J. C., Portalier R., The excC gene of Escherichia coli K-12 required for cell envelope integrity encodes the peptidoglycan-associated lipoprotein (PAL). Mol. Microbiol. 6, 735–742 (1992). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

