Multiplexed Digital Characterization of Misfolded Protein Oligomers via Solid-State Nanopores
- PMID: 37972287
- PMCID: PMC10690769
- DOI: 10.1021/jacs.3c09335
Multiplexed Digital Characterization of Misfolded Protein Oligomers via Solid-State Nanopores
Abstract
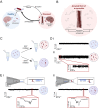
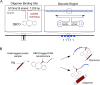
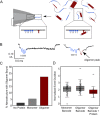
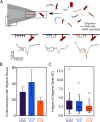
Misfolded protein oligomers are of central importance in both the diagnosis and treatment of Alzheimer's and Parkinson's diseases. However, accurate high-throughput methods to detect and quantify oligomer populations are still needed. We present here a single-molecule approach for the detection and quantification of oligomeric species. The approach is based on the use of solid-state nanopores and multiplexed DNA barcoding to identify and characterize oligomers from multiple samples. We study α-synuclein oligomers in the presence of several small-molecule inhibitors of α-synuclein aggregation as an illustration of the potential applicability of this method to the development of diagnostic and therapeutic methods for Parkinson's disease.
Conflict of interest statement
The authors declare the following competing financial interest(s): S.E.S is funded by Oxford Nanopore Technologies. R.I.H and S.C have been consultants of WaveBreak Therapeutics (formerly Wren Therapeutics). M.C.C is an employee of WaveBreak Therapeutics. M.V is a founder of WaveBreak Therapeutics.
Figures

Similar articles
-
Single-Molecule Detection of α-Synuclein Oligomers in Parkinson's Disease Patients Using Nanopores.ACS Nano. 2023 Nov 28;17(22):22999-23009. doi: 10.1021/acsnano.3c08456. Epub 2023 Nov 10. ACS Nano. 2023. PMID: 37947369 Free PMC article.
-
How specific are the conformation-specific α-synuclein antibodies? Characterization and validation of 16 α-synuclein conformation-specific antibodies using well-characterized preparations of α-synuclein monomers, fibrils and oligomers with distinct structures and morphology.Neurobiol Dis. 2020 Dec;146:105086. doi: 10.1016/j.nbd.2020.105086. Epub 2020 Sep 22. Neurobiol Dis. 2020. PMID: 32971232
-
Rapid Structural, Kinetic, and Immunochemical Analysis of Alpha-Synuclein Oligomers in Solution.Nano Lett. 2020 Nov 11;20(11):8163-8169. doi: 10.1021/acs.nanolett.0c03260. Epub 2020 Oct 20. Nano Lett. 2020. PMID: 33079553 Free PMC article.
-
A glimpse into the structural properties of α-synuclein oligomers.Biofactors. 2024 May-Jun;50(3):439-449. doi: 10.1002/biof.2021. Epub 2023 Dec 8. Biofactors. 2024. PMID: 38063360 Review.
-
Membrane interactions of oligomeric alpha-synuclein: potential role in Parkinson's disease.Curr Protein Pept Sci. 2010 Aug;11(5):334-42. doi: 10.2174/138920310791330659. Curr Protein Pept Sci. 2010. PMID: 20423294 Review.
Cited by
-
Artificial intelligence based advancements in nanomedicine for brain disorder management: an updated narrative review.Front Med (Lausanne). 2025 May 13;12:1599340. doi: 10.3389/fmed.2025.1599340. eCollection 2025. Front Med (Lausanne). 2025. PMID: 40432717 Free PMC article. Review.
-
Characterizing Prion-Like Protein Aggregation: Emerging Nanopore-Based Approaches.Small Methods. 2024 Dec;8(12):e2400058. doi: 10.1002/smtd.202400058. Epub 2024 Apr 21. Small Methods. 2024. PMID: 38644684 Free PMC article. Review.
-
Discovery of potent inhibitors of α-synuclein aggregation using structure-based iterative learning.Nat Chem Biol. 2024 May;20(5):634-645. doi: 10.1038/s41589-024-01580-x. Epub 2024 Apr 17. Nat Chem Biol. 2024. PMID: 38632492 Free PMC article.
-
Detection of protein oligomers with nanopores.Nat Rev Chem. 2025 Apr;9(4):224-240. doi: 10.1038/s41570-025-00694-7. Epub 2025 Mar 5. Nat Rev Chem. 2025. PMID: 40045069 Review.
-
Solid-State Nanopore Real-Time Assay for Monitoring Cas9 Endonuclease Reactivity.ACS Nano. 2025 Jan 28;19(3):3839-3851. doi: 10.1021/acsnano.4c15173. Epub 2025 Jan 15. ACS Nano. 2025. PMID: 39814565 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

