Enhancing histopathological image classification of invasive ductal carcinoma using hybrid harmonization techniques
- PMID: 37973797
- PMCID: PMC10654662
- DOI: 10.1038/s41598-023-46239-0
Enhancing histopathological image classification of invasive ductal carcinoma using hybrid harmonization techniques
Abstract
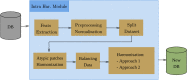
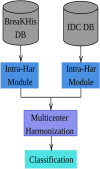
This study aims to develop a robust pipeline for classifying invasive ductal carcinomas and benign tumors in histopathological images, addressing variability within and between centers. We specifically tackle the challenge of detecting atypical data and variability between common clusters within the same database. Our feature engineering-based pipeline comprises a feature extraction step, followed by multiple harmonization techniques to rectify intra- and inter-center batch effects resulting from image acquisition variability and diverse patient clinical characteristics. These harmonization steps facilitate the construction of more robust and efficient models. We assess the proposed pipeline's performance on two public breast cancer databases, BreaKHIS and IDCDB, utilizing recall, precision, and accuracy metrics. Our pipeline outperforms recent models, achieving 90-95% accuracy in classifying benign and malignant tumors. We demonstrate the advantage of harmonization for classifying patches from different databases. Our top model scored 94.7% for IDCDB and 95.2% for BreaKHis, surpassing existing feature engineering-based models (92.1% for IDCDB and 87.7% for BreaKHIS) and attaining comparable performance to deep learning models. The proposed feature-engineering-based pipeline effectively classifies malignant and benign tumors while addressing variability within and between centers through the incorporation of various harmonization techniques. Our findings reveal that harmonizing variabilities between patches from different batches directly impacts the learning and testing performance of classification models. This pipeline has the potential to enhance breast cancer diagnosis and treatment and may be applicable to other diseases.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Classification of benign and malignant subtypes of breast cancer histopathology imaging using hybrid CNN-LSTM based transfer learning.BMC Med Imaging. 2023 Jan 30;23(1):19. doi: 10.1186/s12880-023-00964-0. BMC Med Imaging. 2023. PMID: 36717788 Free PMC article.
-
MuDeRN: Multi-category classification of breast histopathological image using deep residual networks.Artif Intell Med. 2018 Jun;88:14-24. doi: 10.1016/j.artmed.2018.04.005. Epub 2018 Apr 26. Artif Intell Med. 2018. PMID: 29705552
-
Feature Generalization for Breast Cancer Detection in Histopathological Images.Interdiscip Sci. 2022 Jun;14(2):566-581. doi: 10.1007/s12539-022-00515-1. Epub 2022 Apr 28. Interdiscip Sci. 2022. PMID: 35482216
-
RSDCNet: An efficient and lightweight deep learning model for benign and malignant pathology detection in breast cancer.Digit Health. 2025 Apr 16;11:20552076251336286. doi: 10.1177/20552076251336286. eCollection 2025 Jan-Dec. Digit Health. 2025. PMID: 40297351 Free PMC article.
-
Harmonization Strategies in Multicenter MRI-Based Radiomics.J Imaging. 2022 Nov 7;8(11):303. doi: 10.3390/jimaging8110303. J Imaging. 2022. PMID: 36354876 Free PMC article. Review.
Cited by
-
Bilateral Infiltrating Ductal Carcinoma With Adrenal Metastasis: A Rare Case Report.Cureus. 2024 Jul 29;16(7):e65635. doi: 10.7759/cureus.65635. eCollection 2024 Jul. Cureus. 2024. PMID: 39205706 Free PMC article.
-
PND-Net: plant nutrition deficiency and disease classification using graph convolutional network.Sci Rep. 2024 Jul 5;14(1):15537. doi: 10.1038/s41598-024-66543-7. Sci Rep. 2024. PMID: 38969738 Free PMC article.
References
-
- Kitajima K, Miyoshi Y, Sekine T, Takei H, Ito K, Suto A, Kaida H, Ishii K, Daisaki H, Yamakado K. Harmonized pretreatment quantitative volume-based FDG-PET/CT parameters for prognosis of stage I-III breast cancer: Multicenter study. Oncotarget. 2021;12(2):95–105. doi: 10.18632/oncotarget.27851. - DOI - PMC - PubMed
-
- Joann G Elmore, Gary M Longton, Patricia A Carney, Berta M Geller, Tracy Onega, Anna N A Tosteson, Heidi D Nelson, Margaret S Pepe, Kimberly H Allison, Stuart J Schnitt, Frances P O’Malley, Donald L Weaver, “Diagnostic Concordance among Pathologists Interpreting Breast Biopsy Specimens,” JAMA, 2015. doi: 0.1001/jama.2015.1405 - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

