Cryo-EM of Aβ fibrils from mouse models find tg-APPArcSwe fibrils resemble those found in patients with sporadic Alzheimer's disease
- PMID: 37973869
- PMCID: PMC10689242
- DOI: 10.1038/s41593-023-01484-4
Cryo-EM of Aβ fibrils from mouse models find tg-APPArcSwe fibrils resemble those found in patients with sporadic Alzheimer's disease
Abstract
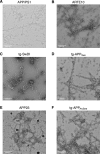
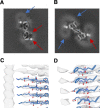
The use of transgenic mice displaying amyloid-β (Aβ) brain pathology has been essential for the preclinical assessment of new treatment strategies for Alzheimer's disease. However, the properties of Aβ in such mice have not been systematically compared to Aβ in the brains of patients with Alzheimer's disease. Here, we determined the structures of nine ex vivo Aβ fibrils from six different mouse models by cryogenic-electron microscopy. We found novel Aβ fibril structures in the APP/PS1, ARTE10 and tg-SwDI models, whereas the human type II filament fold was found in the ARTE10, tg-APPSwe and APP23 models. The tg-APPArcSwe mice showed an Aβ fibril whose structure resembles the human type I filament found in patients with sporadic Alzheimer's disease. A detailed assessment of the Aβ fibril structure is key to the selection of adequate mouse models for the preclinical development of novel plaque-targeting therapeutics and positron emission tomography imaging tracers in Alzheimer's disease.
© 2023. The Author(s).
Conflict of interest statement
L.N.G.N. is on the scientific advisory board and receives a research grant from BioArctic. M.I. is a paid consultant to BioArctic. D.W. is a founder and shareholder of the company Priavoid and a member of its supervisory board. D.W. is co-inventor of patents related to the compound RD2. D.W. is a founder and shareholder of attyloid. D.W. is a member of attyloid’s supervisory board. These had no influence on the interpretation of the data. Benedikt Frieg is now an AstraZeneca employee. All other authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

