Generation of locus coeruleus norepinephrine neurons from human pluripotent stem cells
- PMID: 37974010
- PMCID: PMC11392812
- DOI: 10.1038/s41587-023-01977-4
Generation of locus coeruleus norepinephrine neurons from human pluripotent stem cells
Abstract
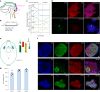
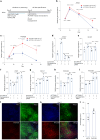
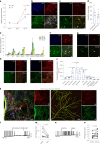
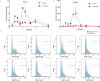
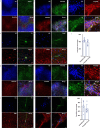
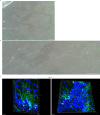
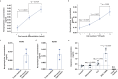
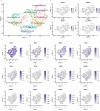
Central norepinephrine (NE) neurons, located mainly in the locus coeruleus (LC), are implicated in diverse psychiatric and neurodegenerative diseases and are an emerging target for drug discovery. To facilitate their study, we developed a method to generate 40-60% human LC-NE neurons from human pluripotent stem cells. The approach depends on our identification of ACTIVIN A in regulating LC-NE transcription factors in dorsal rhombomere 1 (r1) progenitors. In vitro generated human LC-NE neurons display extensive axonal arborization; release and uptake NE; and exhibit pacemaker activity, calcium oscillation and chemoreceptor activity in response to CO2. Single-nucleus RNA sequencing (snRNA-seq) analysis at multiple timepoints confirmed NE cell identity and revealed the differentiation trajectory from hindbrain progenitors to NE neurons via an ASCL1-expressing precursor stage. LC-NE neurons engineered with an NE sensor reliably reported extracellular levels of NE. The availability of functional human LC-NE neurons enables investigation of their roles in psychiatric and neurodegenerative diseases and provides a tool for therapeutics development.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests. S.-C.Z. is a co-founder of BrainXell, Inc.
Figures







References
MeSH terms
Substances
Grants and funding
- R01 MH099587/MH/NIMH NIH HHS/United States
- R24 NS086604/NS/NINDS NIH HHS/United States
- S10 OD018202/OD/NIH HHS/United States
- R01 NS076352/NS/NINDS NIH HHS/United States
- NS096282, NS076352, NS086604/U.S. Department of Health & Human Services | NIH | National Institute of Neurological Disorders and Stroke (NINDS)
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

