Transcriptome sequencing and microRNA-mRNA regulatory network construction in the lens from a Na2SeO3-induced Sprague Dawley rat cataract model
- PMID: 37974089
- PMCID: PMC10652440
- DOI: 10.1186/s12886-023-03202-x
Transcriptome sequencing and microRNA-mRNA regulatory network construction in the lens from a Na2SeO3-induced Sprague Dawley rat cataract model
Abstract
Background: A sight-threatening, cataract is a common degenerative disease of the ocular lens. This study aimed to explore the regulatory mechanism of age-related cataract (ARC) formation and progression.
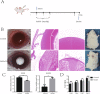
Methods: Cataracts in Sprague Dawley rats were induced by adopting the method that injected selenite subcutaneously in the nape. We performed high-throughput RNA sequencing technology to identify the mRNA and microRNA(miRNA) expression profiles of the capsular membrane of the lens from Na2SeO3-induced and saline-injected Sprague Dawley rats. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were carried out to forecast the regulatory and functional role of mRNAs in cataracts by DAVID and Metascape. The protein-protein interaction(PPI) network of differentially expressed mRNA(DEmRNAs) was built via the STRING. Target miRNAs of hub genes were predicted by miRBD and TargetScan. Furthermore, differentially expressed miRNA(DEmiRNAs) were selected as hub genes' targets, validated by quantitative real-time polymerase chain reaction(qRT-PCR), and a DEmiRNA-DEmRNA regulatory network was constructed via Cytoscape.
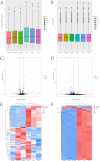
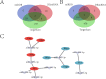
Result: In total, 329 DEmRNAs including 40 upregulated and 289 downregulated genes were identified. Forty seven DEmiRNAs including 29 upregulated and 18 downregulated miRNAs were detected. The DEmRNAs are involved in lens development, visual perception, and aging-related biological processes. A protein-protein interaction network including 274 node genes was constructed to explore the interactions of DEmRNAs. Furthermore, a DEmiRNA-DEmRNA regulatory network related to cataracts was constructed, including 8 hub DEmRNAs, and 8 key DEmiRNAs which were confirmed by qRT-PCR analysis.
Conclusion: We identified several differentially expressed genes and established a miRNA-mRNA-regulated network in a Na2SeO3-induced Sprague Dawley rat cataract model. These results may provide novel insights into the clinical treatment of cataracts, and the hub DEmRNAs and key DEmiRNAs could be potential therapeutic targets for ARC.
Keywords: Cataract; Len; Network; Rat; mRNA; microRNA.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

