PTPRH promotes the progression of non-small cell lung cancer via glycolysis mediated by the PI3K/AKT/mTOR signaling pathway
- PMID: 37974250
- PMCID: PMC10652596
- DOI: 10.1186/s12967-023-04703-5
PTPRH promotes the progression of non-small cell lung cancer via glycolysis mediated by the PI3K/AKT/mTOR signaling pathway
Abstract
Background: The protein tyrosine phosphatase H receptor (PTPRH) is known to regulate the occurrence and development of pancreatic and colorectal cancer. However, its association with glycolysis in non-small cell lung cancer (NSCLC) is still unclear. In this study, we aimed to investigate the relationship between PTPRH expression and glucose metabolism and the underlying mechanism of action.
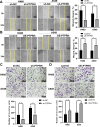
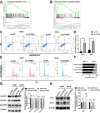
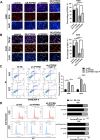
Methods: The expression of PTPRH in NSCLC cells was evaluated by IHC staining, qRT‒PCR and Western blotting. The effect of PTPRH on cell biological behavior was evaluated by colony assays, EdU experiments, Transwell assays, wound healing assays and flow cytometry. Changes in F-18-fluorodeoxyglucose (18F-FDG) uptake and glucose metabolite levels after altering PTPRH expression were detected via a gamma counter and lactic acid tests. The expression of glycolysis-related proteins in NSCLC cells was detected by Western blotting after altering PTPRH expression.
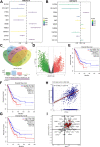
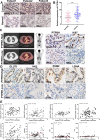
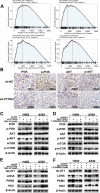
Results: The results showed that PTPRH was highly expressed in clinical patient tissue samples and closely related to tumor diameter and clinical stage. In addition, PTPRH expression was associated with glycometabolism indexes on 18F-FDG positron emission tomography/computed tomography (PET/CT) imaging, the expression level of Ki67 and the expression levels of glycolysis-related proteins. PTPRH altered cell behavior, inhibited apoptosis, and promoted 18F-FDG uptake, lactate production, and the expression of glycolysis-related proteins. In addition, PTPRH modulated the glycometabolism of NSCLC cells via the phosphatidylinositol-3-kinase (PI3K)/protein kinase B (AKT)/mammalian target of rapamycin (mTOR) signaling pathway, as assessed using LY294002 and 740Y-P (an inhibitor and agonist of PI3K, respectively). The same results were validated in vivo using a xenograft tumor model in nude mice. Protein expression levels of PTPRH, glycolysis-related proteins, p-PI3K/PI3K and p-AKT/AKT were measured by IHC staining using a subcutaneous xenograft model in nude mice.
Conclusions: In summary, we report that PTPRH promotes glycolysis, proliferation, migration, and invasion via the PI3K/AKT/mTOR signaling pathway in NSCLC and ultimately promotes tumor progression, which can be regulated by LY294002 and 740Y-P. These results suggest that PTPRH is a potential therapeutic target for NSCLC.
Keywords: Computed tomography; F-18-fluorodeoxyglucose; Glycolysis; Non-small cell lung cancer; Positron emission tomography; Protein tyrosine phosphatase H receptor.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



References
-
- Wood DE, Kazerooni EA, Baum SL, Eapen GA, Ettinger DS, Hou L, Jackman DM, Klippenstein D, Kumar R, Lackner RP, et al. Lung cancer screening, Version 3.2018, NCCN clinical practice guidelines in oncology. J Natl Compr Cancer Netw. 2018;16(4):412–441. doi: 10.6004/jnccn.2018.0020. - DOI - PMC - PubMed
-
- Kim S, Im JH, Kim WK, Choi YJ, Lee JY, Kim SK, Kim SJ, Kwon SW, Kang KW. Enhanced sensitivity of nonsmall cell lung cancer with acquired resistance to epidermal growth factor receptor-tyrosine kinase inhibitors to phenformin: the roles of a metabolic shift to oxidative phosphorylation and redox balance. Oxid Med Cell Longev. 2021;2021:5428364. doi: 10.1155/2021/5428364. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

