Single-Cell Transcriptome Atlas and Regulatory Dynamics in Developing Cotton Anthers
- PMID: 37974530
- PMCID: PMC10797427
- DOI: 10.1002/advs.202304017
Single-Cell Transcriptome Atlas and Regulatory Dynamics in Developing Cotton Anthers
Abstract
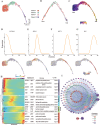
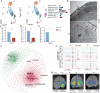
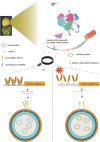
Plant anthers are composed of different specialized cell types with distinct roles in plant reproduction. High temperature (HT) stress causes male sterility, resulting in crop yield reduction. However, the spatial expression atlas and regulatory dynamics during anther development and in response to HT remain largely unknown. Here, the first single-cell transcriptome atlas and chromatin accessibility survey in cotton anther are established, depicting the specific expression and epigenetic landscape of each type of cell in anthers. The reconstruction of meiotic cells, tapetal cells, and middle layer cell developmental trajectories not only identifies novel expressed genes, but also elucidates the precise degradation period of middle layer and reveals a rapid function transition of tapetal cells during the tetrad stage. By applying HT, heterogeneity in HT response is shown among cells of anthers, with tapetal cells responsible for pollen wall synthesis are most sensitive to HT. Specifically, HT shuts down the chromatin accessibility of genes specifically expressed in the tapetal cells responsible for pollen wall synthesis, such as QUARTET 3 (QRT3) and CYTOCHROME P450 703A2 (CYP703A2), resulting in a silent expression of these genes, ultimately leading to abnormal pollen wall and male sterility. Collectively, this study provides substantial information on anthers and provides clues for heat-tolerant crop creation.
Keywords: anthers; high temperature; single-cell RNA sequencing (scRNA-seq); single-cell multi-omics; tapetum.
© 2023 The Authors. Advanced Science published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Single-cell transcriptomic and cell‑type‑specific regulatory networks in Polima temperature-sensitive cytoplasmic male sterility of Brassica napus L.BMC Plant Biol. 2024 Dec 19;24(1):1206. doi: 10.1186/s12870-024-05916-6. BMC Plant Biol. 2024. PMID: 39701979 Free PMC article.
-
High temperature induces male sterility via MYB66-MYB4-Casein kinase I signaling in cotton.Plant Physiol. 2022 Aug 1;189(4):2091-2109. doi: 10.1093/plphys/kiac213. Plant Physiol. 2022. PMID: 35522025 Free PMC article.
-
Rapid Identification of Pollen- and Anther-Specific Genes in Response to High-Temperature Stress Based on Transcriptome Profiling Analysis in Cotton.Int J Mol Sci. 2022 Mar 21;23(6):3378. doi: 10.3390/ijms23063378. Int J Mol Sci. 2022. PMID: 35328797 Free PMC article.
-
Epigenetic Dynamics and Regulation of Plant Male Reproduction.Int J Mol Sci. 2022 Sep 9;23(18):10420. doi: 10.3390/ijms231810420. Int J Mol Sci. 2022. PMID: 36142333 Free PMC article. Review.
-
Thermal stress impacts reproductive development and grain yield in rice.Plant Physiol Biochem. 2017 Jun;115:57-72. doi: 10.1016/j.plaphy.2017.03.011. Epub 2017 Mar 16. Plant Physiol Biochem. 2017. PMID: 28324683 Review.
Cited by
-
Male Germ Cell Specification in Plants.Int J Mol Sci. 2024 Jun 17;25(12):6643. doi: 10.3390/ijms25126643. Int J Mol Sci. 2024. PMID: 38928348 Free PMC article. Review.
-
The complex transcriptional regulation of heat stress response in maize.Stress Biol. 2024 Apr 26;4(1):24. doi: 10.1007/s44154-024-00165-x. Stress Biol. 2024. PMID: 38668992 Free PMC article. Review.
-
Single-cell multiome reveals root hair-specific responses to salt stress.New Phytol. 2025 Jun;246(6):2634-2651. doi: 10.1111/nph.70160. Epub 2025 Apr 23. New Phytol. 2025. PMID: 40269556 Free PMC article.
-
A systematic review of single-cell RNA sequencing applications and insights in Oryza sativa response to environmental stresses.Mol Biol Rep. 2025 Jul 19;52(1):739. doi: 10.1007/s11033-025-10836-1. Mol Biol Rep. 2025. PMID: 40682700 Review.
-
PpMYC2 and PpJAM2/3 antagonistically regulate lignin synthesis to cope with the disease in peach fruit.Plant Biotechnol J. 2025 Sep;23(9):3524-3539. doi: 10.1111/pbi.70177. Epub 2025 Jun 4. Plant Biotechnol J. 2025. PMID: 40468145 Free PMC article.
References
-
- van der Linde K., Walbot V., Curr. Top. Dev. Biol. 2019, 131, 239. - PubMed
-
- Sanders P. M., Bui A. Q., Weterings K., McIntire K. N., Hsu Y.‐C., Lee P. Y., Truong M. T., Beals T. P., Goldberg R. B., Sexual Plant Reproduction 1999, 11, 297.
-
- Zhang D., Luo X., Zhu L., J. Genet. Genomics 2011, 38,379. - PubMed
-
- Walbot V., Egger R. L., Annu. Rev. Plant Biol. 2016, 67, 365. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 2022YFD1200805/National Key R&D Program of China
- 2022hszd0042/Major Project of Hubei Hongshan Laboratory
- 32072024/National Natural Science Foundation of China
- xjnkywdzc-2022001/Key Projects for Crop Traits Formation and Cutting-Edge Technologies in Biological Breeding
- 2023ZKPY003/Fundamental Research Funds for the Central Universities
LinkOut - more resources
Full Text Sources
Medical
